David Mittan-Moreau
Computational Research Scientist
Recent Publications
Recent Publications
Related News
Congratulations to Biosciences Area Director’s Award Recipients
Several Biosciences Area personnel are among the 2024 recipients of Berkeley Lab Director’s Achievement Awards. The program recognizes outstanding contributions by employees to all aspects of Lab activities.
Research Interests
HUNTINGTON’S DISEASE
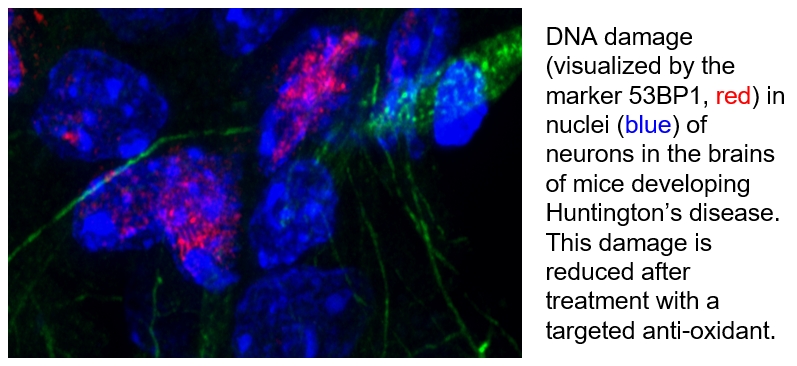 Huntington’s Disease (HD) is one of the longest studied hereditary diseases. The cause has long been known; it has been tracked down to an inherited defect in a single gene (huntingtin). There is also a good understanding of the process leading to this genetic mutation (a trinucleotide repeat expansion). But how this mutated gene leads to the progressive breakdown of nerve cells in the brain is yet to be understood. In our study of the progression of HD, we have recently focused on metabolic effects that HD has in the brain. This altered metabolism has consequences on the oxidative load generated in the brain, which increases along with genetic damage to neural cells. This work might be an important insight into HD neurodegeneration, and provides potential therapeutic strategies to target increased oxidative load in the brain.
Huntington’s Disease (HD) is one of the longest studied hereditary diseases. The cause has long been known; it has been tracked down to an inherited defect in a single gene (huntingtin). There is also a good understanding of the process leading to this genetic mutation (a trinucleotide repeat expansion). But how this mutated gene leads to the progressive breakdown of nerve cells in the brain is yet to be understood. In our study of the progression of HD, we have recently focused on metabolic effects that HD has in the brain. This altered metabolism has consequences on the oxidative load generated in the brain, which increases along with genetic damage to neural cells. This work might be an important insight into HD neurodegeneration, and provides potential therapeutic strategies to target increased oxidative load in the brain.
INFRARED SPECTROSCOPY IMAGING FOR BIOLOGY
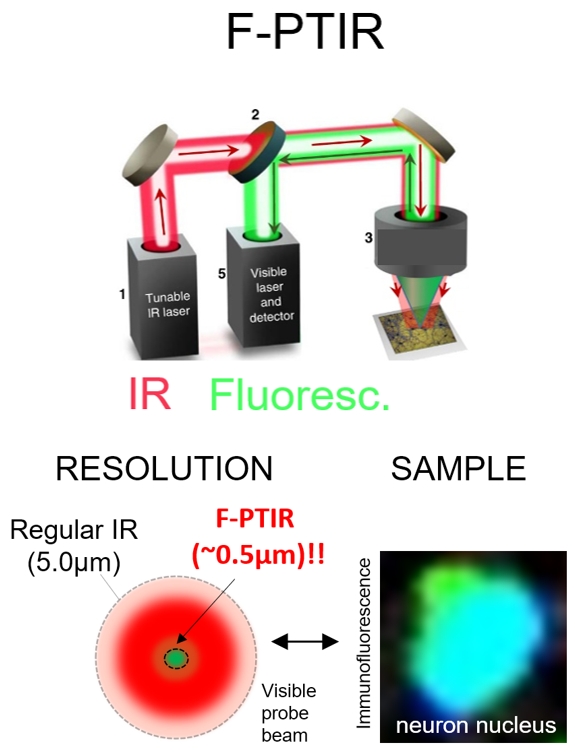 The chemical features common to molecules in living cells have vibrational energies that are within the Infrared (IR) energy range and every biomolecule in a cell has a distinct absorption spectrum in the IR region. This means that if you could image a living cell across the IR spectrum you would, in theory, have a compilation reading of all it’s biomolecules, or a reading of the state of the cell. We are trying to develop and validate IR microscopy of cells and tissue as a tool that could be used for the characterization of disease. We are pursuing the design of new IR imagers based on novel concepts (such as fluorescently based photothermal IR spectroscopy, F-PTIR) that are amenable to imaging biological systems. These systems aid in the high resolution, sensitivity and ability to identify and localize biological features (such as cells in a tissue or microbes in a solution) while acquiring IR absorption spectra of living things.
The chemical features common to molecules in living cells have vibrational energies that are within the Infrared (IR) energy range and every biomolecule in a cell has a distinct absorption spectrum in the IR region. This means that if you could image a living cell across the IR spectrum you would, in theory, have a compilation reading of all it’s biomolecules, or a reading of the state of the cell. We are trying to develop and validate IR microscopy of cells and tissue as a tool that could be used for the characterization of disease. We are pursuing the design of new IR imagers based on novel concepts (such as fluorescently based photothermal IR spectroscopy, F-PTIR) that are amenable to imaging biological systems. These systems aid in the high resolution, sensitivity and ability to identify and localize biological features (such as cells in a tissue or microbes in a solution) while acquiring IR absorption spectra of living things.
HEALTH EFFECTS OF SMOKE EXPOSURES
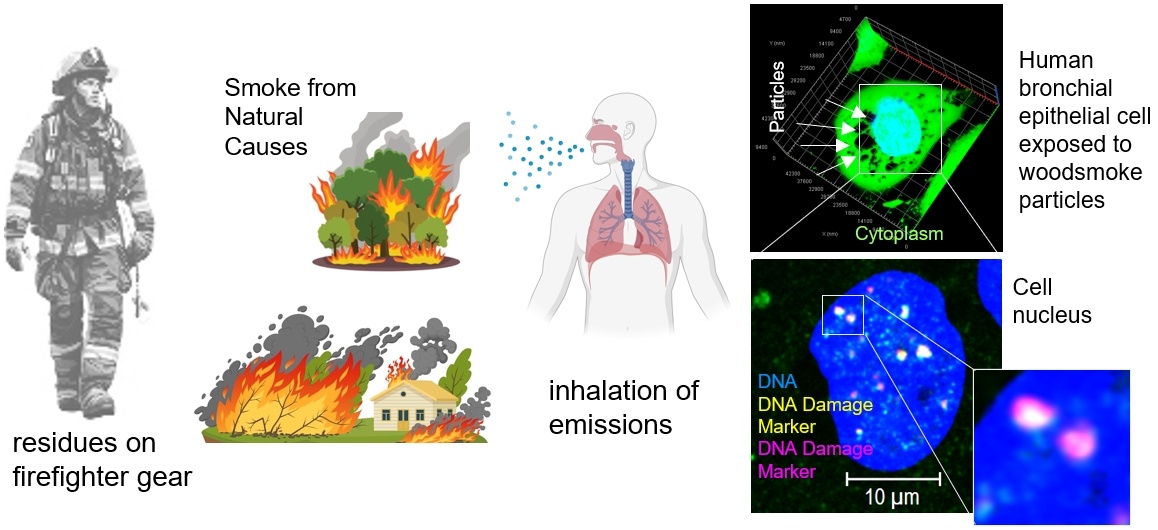 Exposures to smoke from burning vegetation (wildfires, woodland-urban-interface fires (WUI), etc.) is a natural hazard that has been increasing in the United States, which also increases the potential effects on the population. Understanding the long-term health ramifications of these exposures, including cancer and other disease effects, requires both epidemiological/clinical/real world and controlled laboratory studies. We are using laboratory models to study the cellular and tissue effects of wood combustion emissions to understand their carcinogenic and respiratory ramifications. We are also studying the toxicity and variety of exposures to combustion emissions that California firefighters in municipal fire services have. Specifically, we are looking at exposures through the residues that remains on gear after an incident response.
Exposures to smoke from burning vegetation (wildfires, woodland-urban-interface fires (WUI), etc.) is a natural hazard that has been increasing in the United States, which also increases the potential effects on the population. Understanding the long-term health ramifications of these exposures, including cancer and other disease effects, requires both epidemiological/clinical/real world and controlled laboratory studies. We are using laboratory models to study the cellular and tissue effects of wood combustion emissions to understand their carcinogenic and respiratory ramifications. We are also studying the toxicity and variety of exposures to combustion emissions that California firefighters in municipal fire services have. Specifically, we are looking at exposures through the residues that remains on gear after an incident response.
GENETIC DAMAGE
The genome codes the template for life; it stores this vast amount of information in strings of DNA. There are many intricate pathways that maintain the integrity of this information and repair DNA that is damaged. These pathways are a major area of scientific study. We are trying to develop tools to help elucidate the pathways that are activated after damage and understand the implications to the genome. These tools would allow us to target DNA damage (oxidation) to a single genetic site and pick out what proteins are part of the response.
Recent Publications
Related News
Unraveling the Mysteries of DNA Damage in the Brain
A first-of-its-kind study led by Cynthia McMurray and Aris Polyzos in the Molecular Biophysics and Integrated Bioimaging (MBIB) Division integrated cell type– and brain region–specific features of DNA repair in normal brains, setting a benchmark for the field. Their results, recently published in Nature Communications, suggest that DNA damage itself serves as the checkpoint, limiting the accumulation of genomic errors in cells during natural aging.
Biosciences FY25 LDRD Projects
The projects of 23 Biosciences Area scientists and engineers received funding through the FY25 Laboratory Directed Research and Development (LDRD) program.
Biosciences Area FY24 LDRD Projects
The projects of 21 Biosciences Area scientists and engineers received funding through the FY24 Laboratory Directed Research and Development (LDRD) program.

Building: Plant and Environmental Sciences Building, Room 3308
Mail Stop: One Shields Avenue, Davis, CA 95616-8627
Phone: (530) 752-1130
Fax: (530) 752-1552
jmrodrigues@ucdavis.edu
https://soilecogenomicslabs.weebly.com/
https://microbiome.ucdavis.edu/people/jorge-rodrigues
Links
Biography
Jorge “George” Mazza Rodrigues is a professor in the Department of Land, Air, and Water Resources at University of California, Davis. He graduated at the top of his class with a degree in agronomical engineering from the University of Sao Paulo, Brazil, and earned dual PhD degrees from Michigan State University. He was recognized as a UC Davis Faculty Leadership Fellow in 2022 and is currently a member of the First-Generation Faculty Learning Community.
Research Interests
Research in Rodrigues’s laboratory lies at the interface of soil microbial genomics and ecology and focuses on sustainable solutions for biodiversity maintenance in tropical forests and agricultural fields under increased world food demand. He uses a combination of molecular and physiological experimental data to study microbial phylogenetic and functional diversification and their consequences for global biogeochemical cycles. Results from this work have broad implications from basic research in evolutionary biology and ecology to applied aspects, such as conservation policies for natural ecosystems.
Recent Publications
Related News
UC Davis, Berkeley Lab Team up to Advance Green Agriculture
Three University of California, Davis, Faculty Fellows have been awarded $25,000 each to spearhead cross-campus research projects with Berkeley Lab scientists in the field of agricultural decarbonization. With agricultural activities contributing over 10% of the United States’ total greenhouse gas emissions, the sector is a prime target for lowering emissions and addressing the climate crisis. The teams will explore innovative methods to remove and store excess carbon dioxide from the atmosphere—a practice known as carbon sequestration—and minimize energy consumption in crop production.

Building: 91 and 977, Room 091-0450D4 and 977-225
Mail Stop: 977
Phone: MJoachimiak@lbl.gov
http://berkeleybop.github.io/people/marcin-joachimiak/
https://arkinlab.bio/lab-member/marcin-p-joachimiak/
Links
Biography
Marcin Joachimiak is a staff researcher in the Environmental Genomics and Systems Biology Division at Lawrence Berkeley National Laboratory (Berkeley Lab). His current research is performed as part of the DOE Systems Biology Knowledgebase (KBase), the NIH Bridge2AI project, as well as a Berkeley Lab-funded project for predicting microbial growth conditions. He has twenty-five years of experience and a publishing record (H-index 30) in developing and evaluating machine learning algorithms, methods for functional genomics, bioinformatics data science, knowledge modeling, and computational systems design for computational biology. For the past 17 years he has been a member of large distributed microbial and biomedical projects with either a large computational component interfacing with experimentalists (DOE ESPP, DOE ENIGMA Science Focus Area) or which are computational systems centered around modeling and inferring knowledge (KBase, Monarch Initiative, NCATS Biomedical Data Translator, NIH Phenomics First, NIH Bridge2AI). He has led or currently leads team efforts in functional genomics analysis (ENIGMA, KBase, NCATS Translator), knowledge modeling (KBase, Bridge2AI), and machine learning algorithm development (ENIGMA, KBase, NCATS Translator).
Research Interests
Marcin Joachimiak is a computational biologist dedicated to the integration of microbial biology with machine learning and semantic technologies. At the forefront of my research interests is the development of machine learning techniques, including graph learning and biclustering, to unravel the complex relationships in microbiology and metagenome data. Additionally, by leveraging semantic technologies and ontologies, I aim to enhance machine learning, data science, and knowledge graph construction for microbes and microbiomes – a more nuanced and dynamic framework for studying microbial life. As an example, I am developing advanced models for growth prediction and culturing medium optimization using knowledge graphs and graph learning, including tailoring to the unique environment characteristics of different microbiomes. This research has the potential to improve our ability to characterize microbes and develop new utilitarian purposes such as in biomanufacturing, environmental remediation, and human health. Via synthesis of these computational methods and harmonized metagenome and microbiological data, new possibilities emerge in managing and manipulating microbes and microbiomes.
Recent Publications
Related News
Biosciences FY25 LDRD Projects
The projects of 23 Biosciences Area scientists and engineers received funding through the FY25 Laboratory Directed Research and Development (LDRD) program.
Biosciences Area FY24 LDRD Projects
The projects of 21 Biosciences Area scientists and engineers received funding through the FY24 Laboratory Directed Research and Development (LDRD) program.
Biomedical Data Translator Consortium Reports Progress in Pair of Publications
In a pair of recently published papers, members of the Biomedical Data Translator Consortium detailed new features, functionality, and applications of the Translator system and its underlying data model, the Biolink Model.
Biography
Bronwyn Lucas is an assistant professor of biochemistry, biophysics and structural biology in the Department of Molecular and Cell Biology, as well as an assistant professor in the Center for Computational Biology at the University of California, Berkeley.
Research Interests
The Lucas Lab develops methods to visualize molecular structures in cells using focused ion beam (FIB-) milling and cryogenic electron microscopy (cryo-EM). They adapt structural biology techniques as tools for understanding the molecular mechanisms of RNA folding and RNA-protein complex formation within the native cellular environment.
Recent Publications
No publications are available at this time.

Building: 978, Room 4121E
Mail Stop: 978
Phone: (305) 323-8426
mthompson@lbl.gov
https://www.jbei.org/person/mitchell-thompson-2/
Research Interests
- Functional Genomics
- Synthetic Biology
- Microbiology
- Plant-Microbe Interactions
Recent Publications
Research Interests
My research interest has been focused on how genetic and/or epigenetic variation is translated to complex phenotypic variation. I have been combining functional genomics and epigenomics as well as synthetic biology tools for my research.
Research areas are:
- Microbiology
- Plant Biology
- Crop and Pasture Production
- Industrial Biotechnology
Recent Publications

Building: 80A, Room 0100A
Mail Stop: 80R0114
Phone: (510) 486-7765
yangha@lbl.gov
https://bcsb.als.lbl.gov/
Research Interests
Protein crystallography
Recent Publications
Research Interests
I am interested in understanding metalloproteins with an active catalytic site with a focus on photosynthetic proteins, as well as the development of biomimetic catalytic systems relevant to the conversion and storage of renewable energies.
Keywords:
photosynthesis; protein function; light induced charge separation; proton coupled electron transfer; catalytic reactions; protein binding sites of substrates and cofactors; redox potentials; solar fuels; and design of catalytic systems.
Methods:
infrared spectroscopy; time-resolved spectroscopy; fluorescence methods; serial femtosecond crystallography; X-ray absorption and emission spectroscopy; and setup development.
Recent Publications
Related News
New Method Reveals Evolution of a Copper Catalyst
A new method devised by researchers from SLAC National Accelerator Laboratory and Berkeley Lab enables observation of the few copper atoms that actively participate in catalytic reactions while ignoring the rest. The technique can be applied to studying a wide range of energy technologies.
Congratulations to Biosciences Area Director’s Award Recipients
Several Biosciences Area personnel are among the 2024 recipients of Berkeley Lab Director’s Achievement Awards. The program recognizes outstanding contributions by employees to all aspects of Lab activities.
Research Interests
My research interests focus on the development of advanced cellular models to elucidate the mechanisms underlying disease and infection pathogenesis.
Biography
Cat Adams is a fungal ecologist interested in how chemistry such as toxins, light, or fragrances drive interactions between fungi and other living things. She earned her BS from the University of Washington in Biology with emphasis on Ecology and Evolution. For her Master’s at Harvard she studied how fungal seed pathogens of wild chili peppers evolved tolerance to spice. Her PhD at Cal examined the role of toxins in the invasion success of the death cap mushroom, Amanita phalloides. Her postdoctoral research examined how fragrances made by a plant root-associated fungus impact their friendly neighborhood bacteria. Cat is excited to join the Northen lab and learn more about mycorrhizal fungi!
Cat also enjoys embroidery, acrylic pour painting, and metal music.
Research Interests
Chemistry, ecology, fungi, mushrooms, signalling, soil, and toxins.











