
Led by EGSB, EcoFAB involves close collaborations with researchers at the DOE Joint Genome Institute and in the Earth and Environmental Sciences Area. A cross-functional team of biologists, geologists, and ecologists from Berkeley Lab will provide critical new insights into ecosystem processes through the creation of controlled model ecosystems in which microorganisms and host responses can be monitored in response to additional or changing variables.

EcoPODs are enclosed environments that allow direct and intensive monitoring and manipulation of replicated plant-soil-microbe-atmosphere interactions over the complete plant life cycle. These several-cubic-meter, “pilot-scale” ecosystems are designed to bridge the gap between small, lab-scale experiments that are not large enough to mimic the environment and field-scale experiments that cannot be carefully controlled. The goal is to develop cross-disciplinary partnerships that will use the EcoPODs to develop testable ecosystem models in topics that include carbon cycling and secure biosystems design.

The mission of ENIGMA is to support the development of laboratory and computational tools that link the molecular functions within individual microbes to the integrated activities of microbial communities as they interact with their environment.

The GO knowledgebase is the world’s largest source of information on gene functions. Both human-readable and machine-readable, this knowledge is foundational for computational analysis of large-scale molecular biology and genetics experiments in biomedical research. GO is a multi-PI collaboration among Berkeley Lab, the University of Southern California, Stanford University, and the California Institute of Technology, in addition to a very broad consortium.

KBase is an open platform for comparative functional genomics and systems biology for microbes, plants and their communities, and for sharing results and methods with other scientists. Berkeley Lab is the lead institution in a partnership comprised of Argonne, Brookhaven, and Oak Ridge National Laboratories.

Collaborative, coordinated, and integrated, m-CAFES is a mission-driven program to interrogate the function of soil microbiomes with critical implications for carbon cycling and sequestration, nutrient availability and plant productivity in natural and managed ecosystems. The project targets molecular mechanisms governing carbon and nutrient transformation in soil, with a focus on microbial metabolic networks.
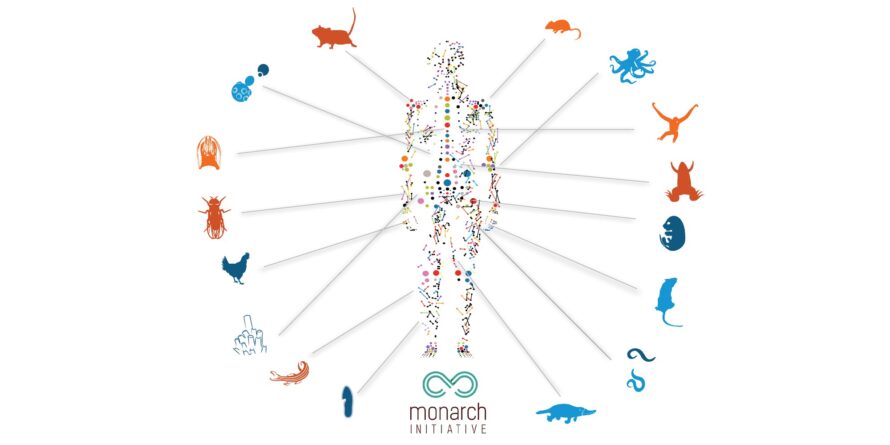
The Monarch Initiative is an integrative data and analytic platform connecting phenotypes to genotypes across species, bridging basic and applied research with semantics-based analysis. Berkeley Lab co-leads this project with the University of Colorado and the Jackson Laboratory.

A multi-national laboratory partnership led by Berkeley Lab, NMDC seeks to address fundamental roadblocks in microbiome data science and gaps in transdisciplinary collaboration. Its two strategic priorities—infrastructure and engagement—and future activities support the long-term vision of empowering the research community to more effectively harness microbiome data.
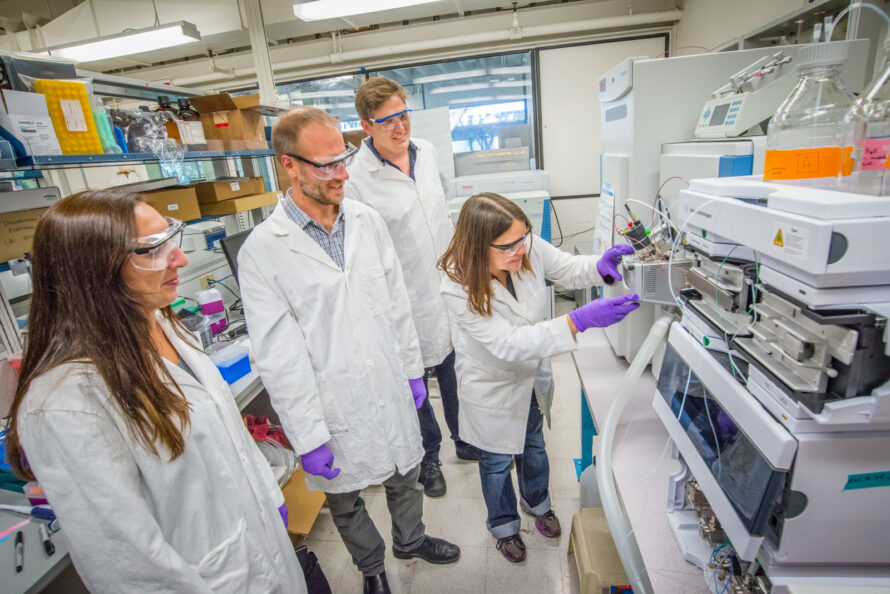
The OpenMSI project facilitates researchers’ use of mass spectrometry imaging (MSI) by providing a web-based gateway for management and storage of MSI data, visualization of the hyper-dimensional contents of the data, and statistical analysis. OpenMSI uses the advanced high-performance compute resources of the National Energy Research Scientific Computing Center (NERSC) to make advanced mass spectrometry imaging accessible.
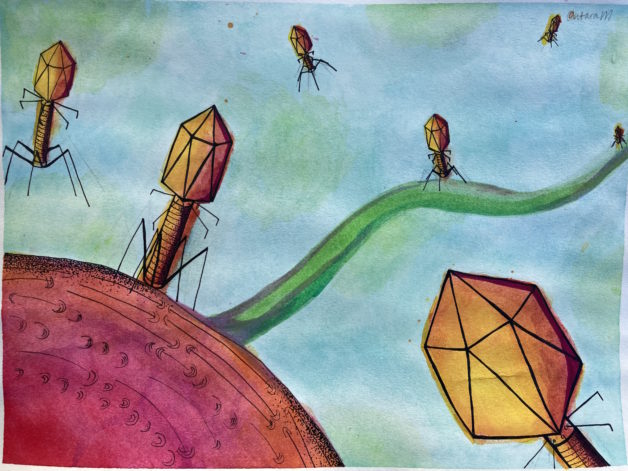
The Phage Foundry is a high-throughput platform for rapid design and development of countermeasures to combat emerging drug-resistant pathogens.

A multi-institution collaboration led by Berkeley Lab, RESTOR-C seeks to understand and improve how plants and microbes remove carbon dioxide from the atmosphere and stably store it for more than 100 years in the soil.



