By understanding biological processes vital to health, we will expedite solutions and resilience by fueling the foundational scientific discovery pipeline. We aim to identify, describe, and understand environmental impacts on human biology, including the human microbiome and virome. We will enable and empower national preparedness against emerging illnesses and pandemics through the development of diagnostics, treatments, and mitigations for biopreparedness.

Structural biology and imaging
Using cryo-electron microscopy, X-ray crystallography, and X-ray tomography to reveal insights into biological structures such as enzymes, antibodies, and therapeutic targets.
DNA repair
Investigating the cellular mechanisms responsible for repairing breaks and defects in DNA strands — processes that, when disrupted, can cause cancer and other diseases.
Genomics
Building on our contributions to the Human Genome Project, our ongoing genomics work seeks to understand how gene coding and noncoding regulatory DNA regions give rise to cellular functions.
Thirdhand smoke
Researching the health effects that chemicals from tobacco products have on those exposed to contaminated surfaces and objects.
Cancer research
Using human cells and animal models to study the genetic, epigenetic, cellular, and environmental factors that contribute to cancer.
Neurodegenerative disease research
Combining cell and tissue imaging and neuroscience to improve our understanding of the anatomical, biochemical, and neurochemical underpinnings of neurodegenerative diseases.
Bioresilience
Developing advanced models to elucidate the mechanisms underlying disease and infection pathogenesis.
Biopreparedness
Applying expertise in bioinformatics to generate high-quality, curated reference datasets for comparing to emerging data; developing biosensors for the identification of emerging pathogens and toxins; and utilizing basic synthetic biology foundations to enable decontamination, phages, vaccines, adjuvants, and other treatment approaches and delivery mechanisms.
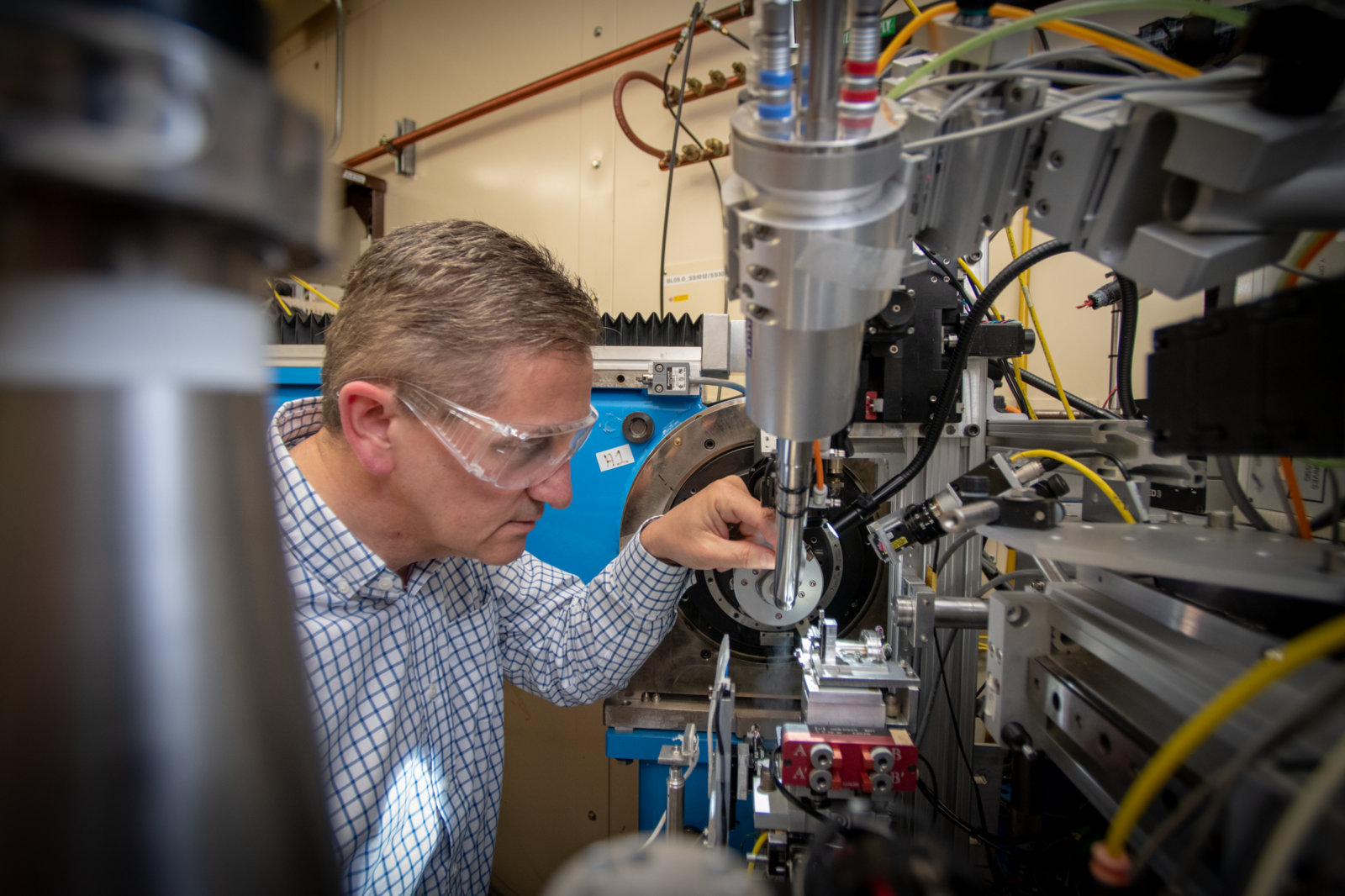
A collective of X-ray beamlines at the Advanced Light Source (ALS) that are dedicated to crystallography and small-angle scattering for cutting-edge structural biology investigations.

BBDS uses computational technology to facilitate and nurture data-intensive biomedical science focused on microbiomes and the genetic and environmental drivers of disease.

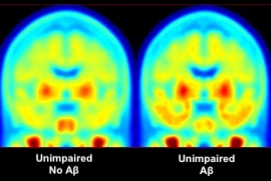
Studying normal brain aging processes and neurodegenerative disease pathology using positron emission tomography (PET), structural magnetic resonance imaging (MRI), functional magnetic resonance imaging (fMRI), neuropsychology, and cognitive neuroscience.

The BSISB program brings 10 years of experience in technology fusion to create and provide new methods that transform infrared spectral imaging, enabling biochemical and structural solutions on complex biological systems.

Our researchers have more than 20 years of experience in the characterization of tobacco smoke residue and its consequences for indoor air quality and human health.

The CCI develops technology and software to improve the speed and ease of molecular structure solving using X-ray crystallography, neutron diffraction, and electron cryo-microscopy (cryo-EM) datasets.

The Gene Ontology (GO) knowledgebase is the world’s largest source of information on gene functions. Both human-readable and machine-readable, this knowledge is foundational for computational analysis of large-scale molecular biology and genetics experiments in biomedical research.
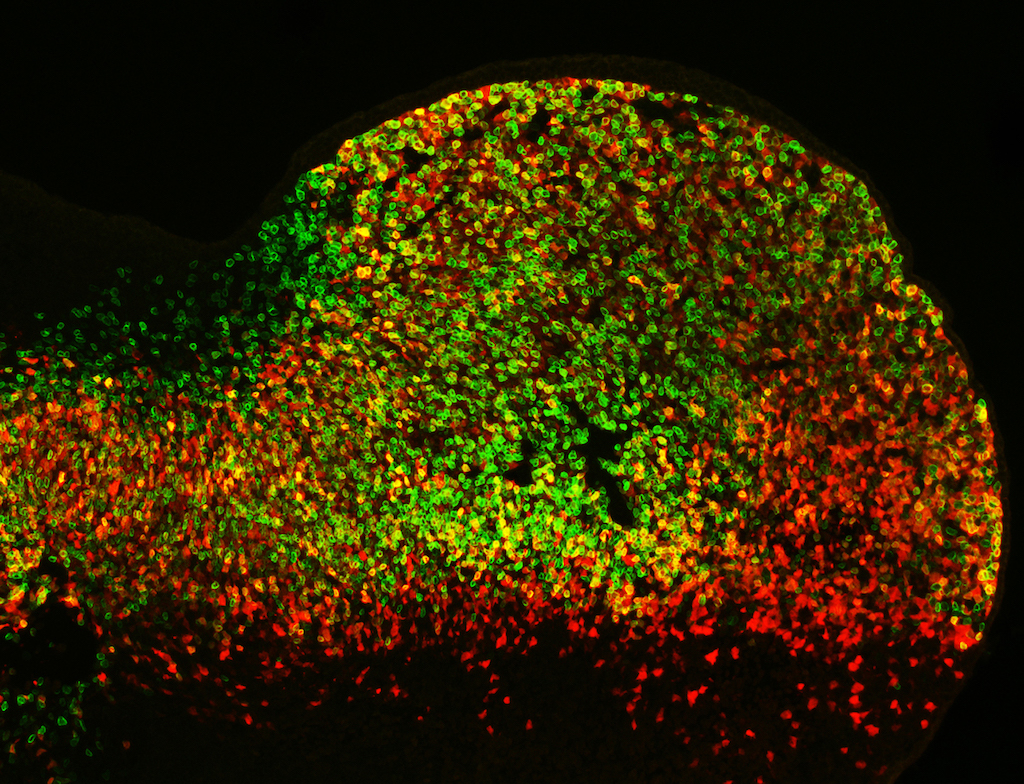
This research group studies DNA function within mammalian genomes and how it goes awry in disease, with a focus on noncoding DNA and epigenomics.

Our soft X-ray tomography capabilities generate detailed 3D maps of the structures inside cells, down to individual organelles.

SBDR is a multi-investigator, multi-institutional, interdisciplinary effort optimized to meet the extreme challenges of characterizing transient complexes and conformations acting in DNA repair (DR) responses. The program will integrate knowledge of DR proteins and pathways by analyzing keystone complexes functioning as regulatory nodes in complex cellular responses to DNA damage.

Antimicrobial-resistant (AMR) infections will require novel antibiotics. Bacteriophages (phages) – viruses that target bacteria – offer a powerful alternative approach to combat AMR bacterial infections. The Phage Foundry serves as an open platform integrating many aspects of phage research to enable rapid development of targeted phage-based therapeutics against AMR pathogens.

Taskforce 5 is a multi-institutional team of scientists creating the infrastructure and capabilities to transition from being bio-responsive to being bio-prepared. They are developing and testing three plug-and-play integrated DOE bio-imaging pipelines for rapid processing of therapeutic, diagnostic, and vaccine countermeasures against emerging biothreats.
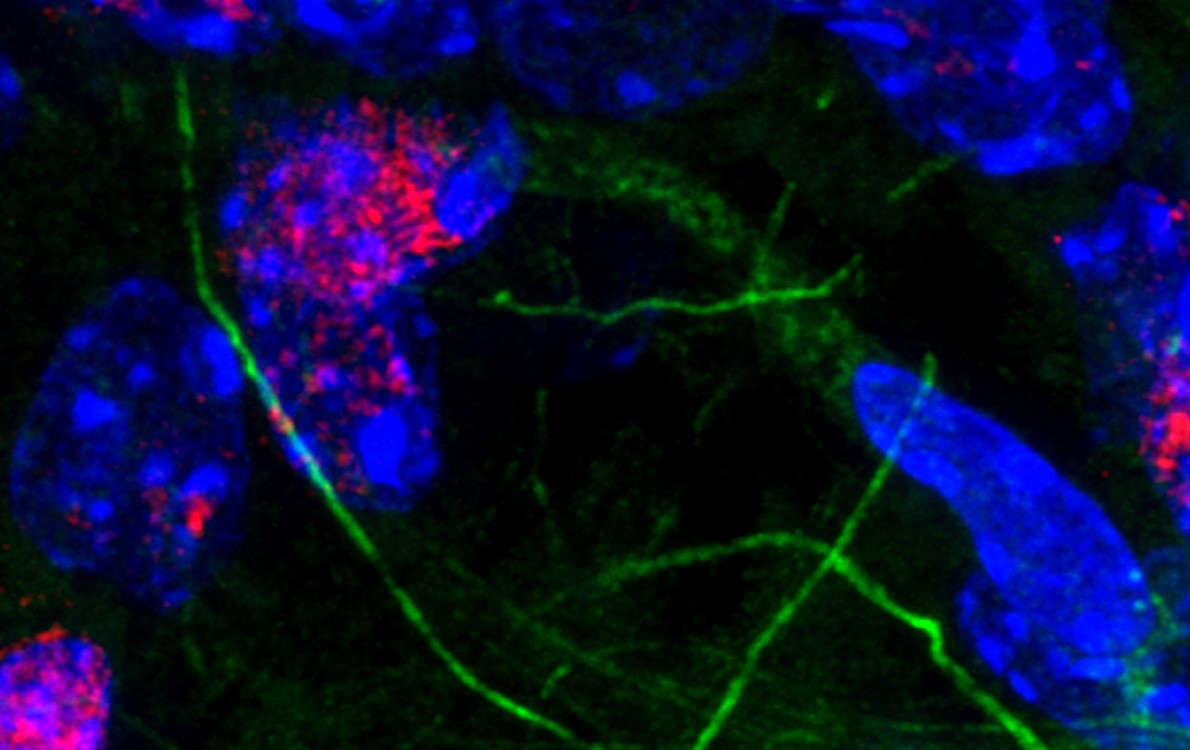

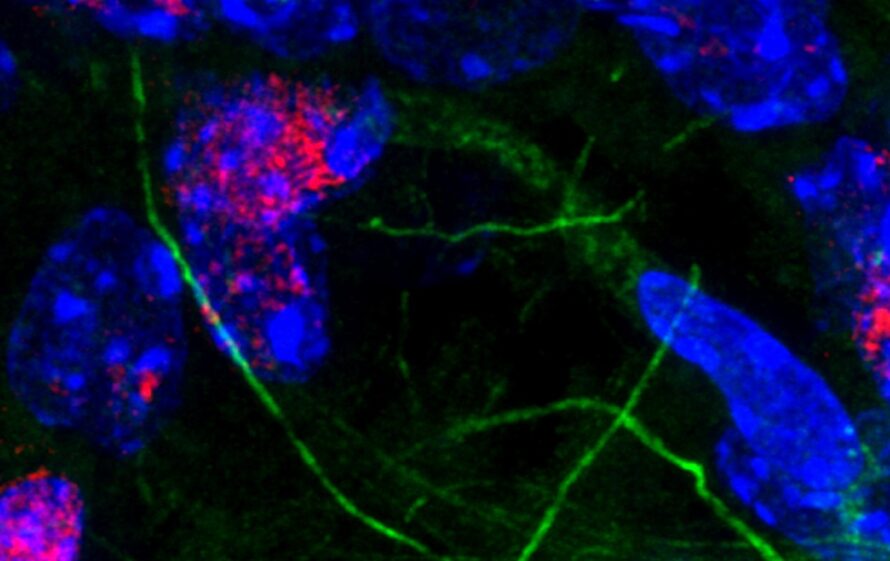
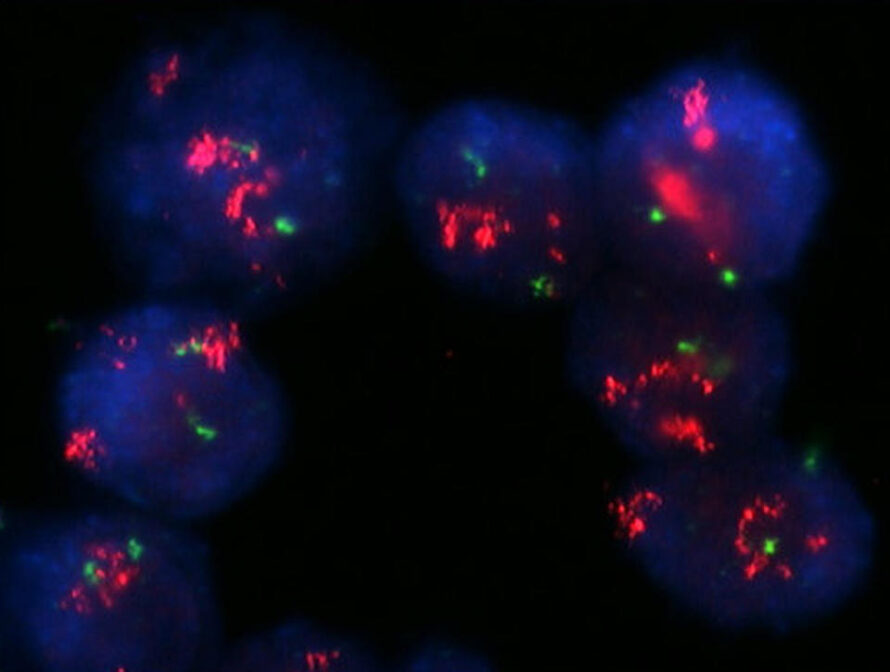
A first-of-its-kind study led by researchers in the Molecular Biophysics and Integrated Bioimaging (MBIB) Division integrated cell type–and brain region–specific features of DNA repair in normal brains, setting a benchmark for the field. Their results suggest that DNA damage itself serves as the checkpoint, limiting the accumulation of genomic errors in cells during natural aging.