The SIBYLS beamline at the Advanced Light Source was used to characterize proteins dreamt up by a reinforcement learning algorithm. The algorithm, developed by researchers in David Baker’s lab at the University of Washington, is powered by the machine learning strategy behind computer programs capable of defeating top human players at board games like chess and go.
Published April 21 in Science, the advance could create a pathway to greater control when designing therapeutic proteins, vaccines, and other molecules. One protein made with the new technique was found to effectively stimulate the formation of new blood vessels in human stem cells and generate a protective immune response in mice.
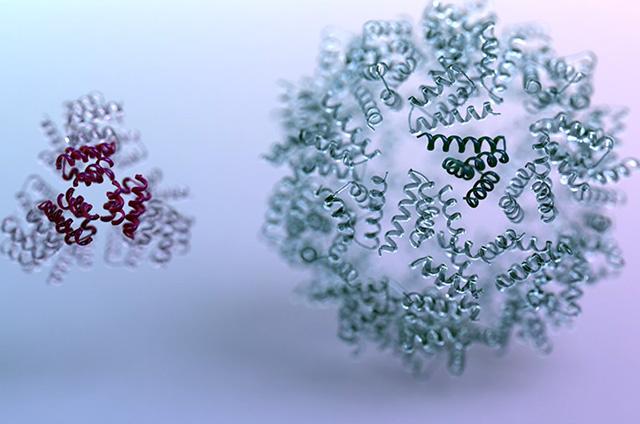
Previous “bottom-up” protein design approaches, which involve assembling small molecular building blocks into larger molecules, have failed to match evolution’s capability of producing proteins that fit together like puzzle pieces in biologically functional ways. This new process, which centers on directing a reinforcement learning algorithm to build proteins from the “top-down” while optimizing for custom architectural and geometric constraints, has the potential to transform the types of molecules scientists can build.
When the researchers brought their AI-designed blueprints into the lab and assembled them into proteins, they found the new design approach to be not only customizable, but also accurate. Small-angle X-ray scattering (SAXS) data collected on the SIBYLS beamline with help from a team led by Molecular Biophysics and Integrated Bioimaging staff scientist Greg Hura revealed the software’s ability to accurately predict molecular architecture. SAXS analysis allowed the researchers to confirm their designs formed as expected in solution.
The accuracy and flexibility of this reinforcement learning-powered method opens the door for researchers to create viable therapeutic molecules with a degree of precise customization not possible with prior approaches.