Molecular Biophysics and Integrated Bioimaging (MBIB) Division faculty scientists James Fraser and James Holton were part of a team that demonstrated that a natural language processing AI can design novel proteins that function as well as naturally occurring ones. This advance could energize the 50-year-old field of protein engineering by speeding the development of new proteins that can be used for almost anything from therapeutics to degrading plastic.
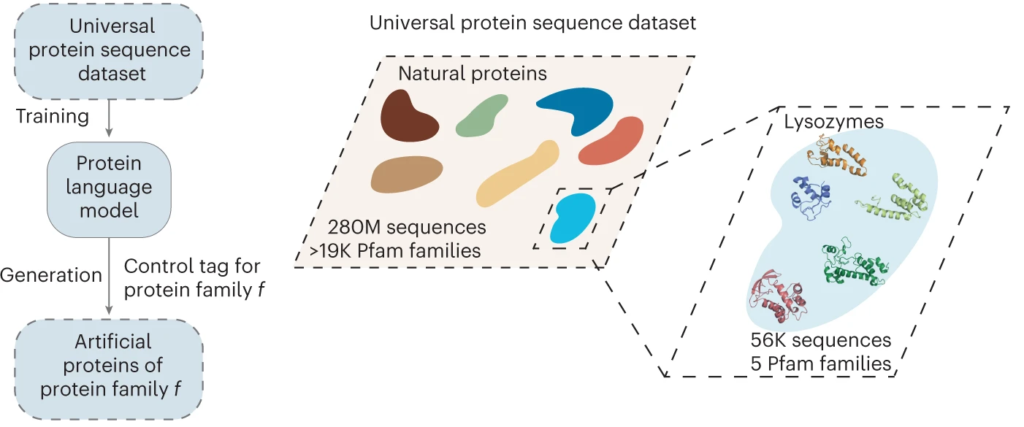
The AI, called ProGen, was originally developed by Salesforce Research to read and compose text. In this case though, instead of learning semantics, parts of speech, and grammar of the English language, the AI learned the language of biology: the genetic code. That enabled it to use next-token prediction to string together amino acid sequences to design novel proteins—specifically, enzymes called lysozymes.
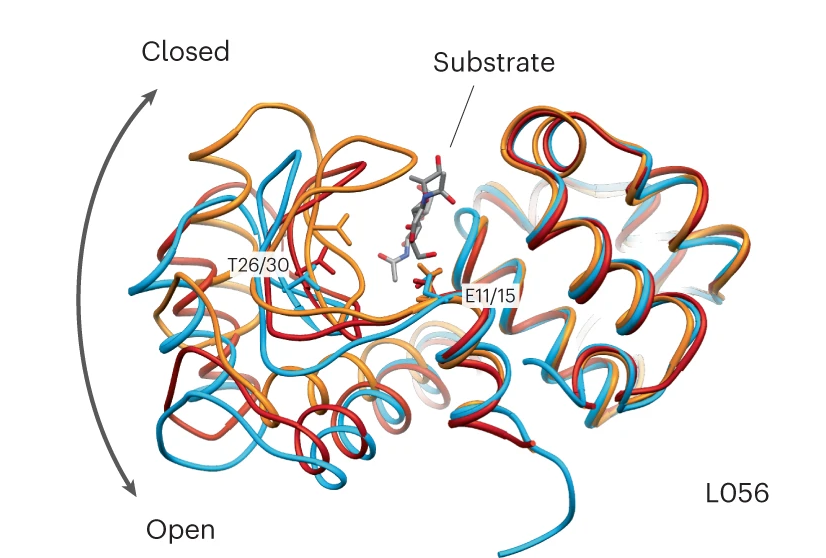
Of the million designs generated by the model, the research team selected 100 to be screened in vitro by Tierra Biosciences, based on how closely they resembled the sequences of natural proteins. Eric Greene, a postdoctoral researcher in Fraser’s UC San Francisco lab, expressed five of the artificial enzymes in cells and compared their activity to an enzyme found in the whites of chicken eggs, known as hen egg white lysozyme (HEWL). Two of the AI-designed enzymes showed activity comparable to HEWL.
Measured with X-ray crystallography at the Advanced Light Source’s beamline 8.3.1, which Holton runs, the atomic structures of the artificial enzymes looked just as they should, although the sequences were like nothing seen before.
Read more from UC San Francisco.
Read more in ALS News.