New Berkeley Lab discovery provides target for developing strategies to control microbial behavior
When bacteria are put in different environments, such as one that is more acidic or anaerobic, their genes start to adapt remarkably quickly. They’re able to do so because the proteins making up their chromosome can pack and unpack rapidly. Now, a Berkeley Lab-led team of researchers has been able to capture this process at the molecular level using advanced imaging techniques, a discovery that could eventually enable scientists to develop strategies to control microbial behavior.
The researchers used multiple high-powered X-ray techniques at Berkeley Lab’s Advanced Light Source (ALS), a Department of Energy Office of Science user facility, to image the process in E. coli bacteria at the micro-, meso-, and nanoscales. The imaging technique they developed enabled them to visualize the bacteria’s chromosome at higher resolutions than ever before, and without the need for labeling, which slows down the process but is required by most other techniques. Their study was published recently in the journal Nature Communications.
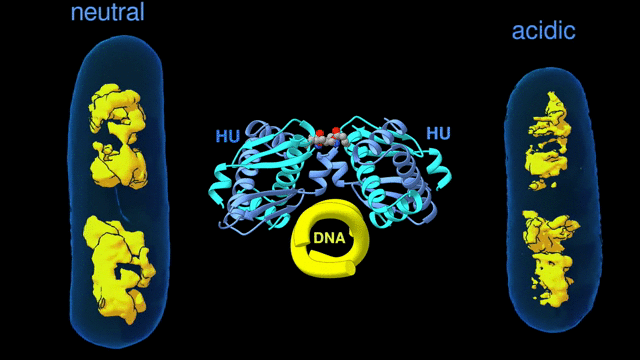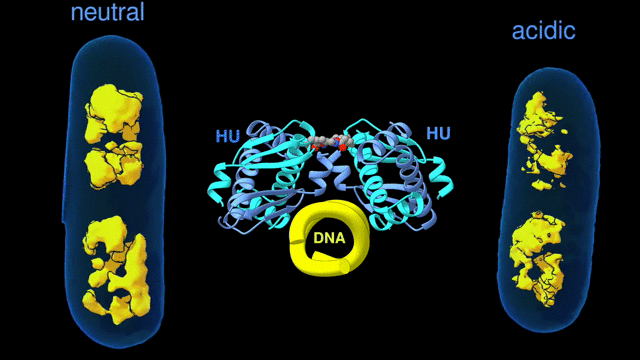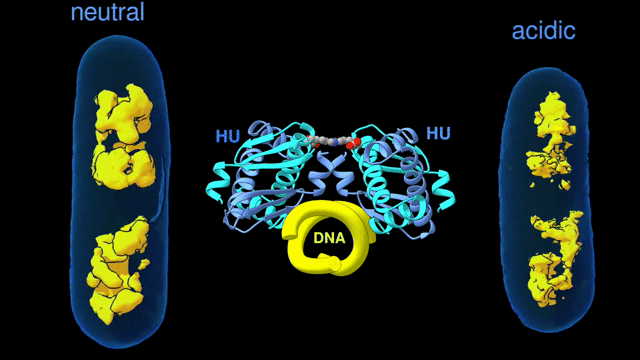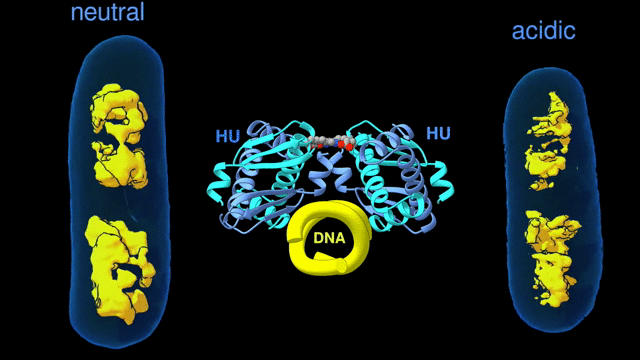
Change in the architecture of the bacterial chromosome during the adaptation to an acidic environment is controlled by the DNA binding protein called HU and its interaction with DNA. (Credit: Michal Hammel/Berkeley Lab)
“We now understand how the packing of the DNA is controlled by the DNA binding proteins, called HU, interacting with each other,” said Michal Hammel, the corresponding author of the paper and a research scientist in Berkeley Lab’s Molecular Biophysics and Integrated Bioimaging (MBIB) Division. “So now we can try to figure out how to control it, how to inhibit or accelerate the packing of the DNA. If you can change packing of DNA, you can change bacterial behavior; then you can start developing alternative approaches to fighting bacterial infections.”
The team included Carolyn Larabell, MBIB faculty scientist and director of the National Center for X-Ray Tomography at the ALS, and the lead authors were Soumya Govinda Remesh and Subhash Verma of the National Cancer Institute.
This Science Snapshot was published on the Berkeley Lab News Center.



