Michal Hammel
Biophysicist Staff Scientist

Building: 6, Room 2104
Mail Stop: 6R2100
Phone: 510 457 6317
MHammel@lbl.gov
https://bl1231.als.lbl.gov/
Links
Research Interests
My early scientific career began with the characterization of drug delivery in Low-Density Lipoprotein and mastering solution X-ray scattering (SAXS) techniques at the Austrian Academy of Sciences in Graz, Austria. I have been bridging crystallography with solution scattering since 2000. My early developments in integrative structural biology were later awarded Erwin Schrödinger fellowship to pursue my postdoctoral research in Marseille at Centre National de la Recherche Scientifique. Here I developed an ensemble modeling approach to investigate flexible and dynamic macromolecules by combining solution scattering, crystallography, and molecular dynamics. This early innovation becomes the groundwork for our current worldwide used FoXS software package.
Besides developing novel SAXS methods, my research focuses on visualizing the large megadalton biological assemblies that included the cellulosome, human complement, and non-homologous end-joining. Characterizing these dynamic molecules impacts medical research, like defining the allosteric mechanism for complement inhibition by the Staphylococcus aureus and discovering the extended grooved scaffold for DNA ligation in DNA break repair.
My effort as the beamline scientist at the SIBYLS beamline at the Advanced Light Source (ALS) makes the SAXS technique accessible to more investigators and pushes SAXS onto a new paradigm. We have been the first to create a high throughput SAXS pipeline and established a mail-in program at SIBYLS to collect SAXS data for scientists from all around the world.
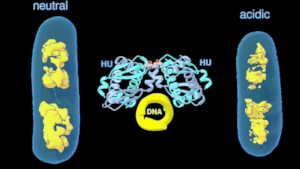
As a research scientist at the LBNL, my research focuses on the modulation of transcription by DNA control elements. Using an altered bacterial histone-like protein, we show that reorganization of the bacterial chromatin can dynamically modulate the cellular transcription pattern. We anticipate making histone-like protein interactions an attractive target for controlling pathogenesis and microbial systems in general with this ongoing research.
Change in the architecture of the bacterial chromosome during the adaptation to an acidic environment is controlled by the DNA binding protein called HU and its interaction with DNA. (Credit: Michal Hammel/Berkeley Lab)
I’m co-directing Structural Cell Biology Core of the NCI-funded Structural Cell Biology of DNA Repair (SBDR) group.
Recent Publications
Related News
SIBYLS Team Recognized with Innovative Instrumentation Award
The Structurally Integrated BiologY for the Life Sciences (SIBYLS) team received the 2025 Klaus Halbach Award for Innovative Instrumentation at the 2025 Advanced Light Source (ALS) User Meeting in August.
A Fast Track for Visualizing RNA Structures
Scientists have combined multiple AI tools into a single streamlined process that predicts the atomic structures of RNA molecules.
Researchers Leverage SAXS to Understand Aspect of Microbial Metabolism
A team of Molecular Biophysics and Integrated Bioimaging Division researchers used synchrotron technology unique to the Advanced Light Source (ALS) at Berkeley Lab to probe the conformational states behind electron bifurcation.