The last common ancestor of humans and rodents lived more than 80 million years ago, and billions of changes in their respective DNA sequences have occurred over this vast timespan. Yet, intriguingly, there are a few hundred stretches of DNA in our genome that are still identical to the corresponding sequences in mice and rats. Generally, sequence conservation between distantly related species is an indication that the function the DNA serves is vitally important and highly sensitive to mutations. For example, most DNA sequences that encode proteins show at least moderate conservation in evolution. However, more than two-thirds of the “ultraconserved” sequences shared by humans and rodents are outside of protein-coding genes, raising the question of what led to their extreme level of conservation.
An initial hint at the function of ultraconserved sequences was provided in 2006 when Berkeley Lab researchers showed that many of them are transcriptional enhancers—molecular switches that turn on the expression of important genes at key times during embryonic development. While this discovery provided a possible explanation as to why the sequences are strongly conserved in evolution, it also predicted that their disruption by deletion or mutation should cause misregulation of genes and impacts on health, general fitness, or fertility. So it came as a surprise when the same team demonstrated a year later that ultraconserved sequences that control gene expression in the brain during development could be removed from the mouse genome, seemingly without negative effects.
Postulating that perhaps deletion of ultraconserved enhancers led to very subtle negative effects on health, members of the Mammalian Functional Genomics Laboratory in Biosciences’ Environmental Genomics and Systems Biology (EGSB) Division deleted additional ultraconserved enhancers in a 2018 follow-up study. Once again, the animals remained viable and fertile, but closer examination of their brains—guided by knowledge about the regions in which the enhancers are normally active—revealed structural abnormalities. While the work clearly demonstrated a function that provided a general explanation for evolutionary conservation of ultraconservered sequences, it still did not explain why they were so perfectly conserved.

From left: Diane Dickel, Len Pennacchio, and Axel Visel of the Mammalian Functional Genomics Lab.
In an effort to resolve this apparent paradox, the team—led by co-PIs Diane Dickel, Len Pennacchio, and Axel Visel—recently explored the possibility that the regulatory functions of ultraconserved elements are exquisitely sensitive to mutations, which would explain why they remained unchanged for tens of millions of years. “Given their perfect conservation, our working hypothesis was that these particular enhancers might be ‘ultrasensitive’ to even the smallest changes in their sequence,” said Dickel, a staff scientist in EGSB.
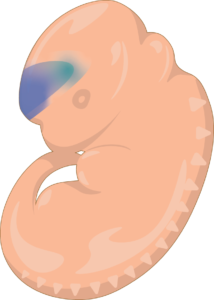
Blue staining indicates enhancer activity in the developing brain.
To test their idea, the researchers selected 23 ultraconserved enhancers and produced artificially mutated versions of each where 2%, 5%, or 20% of the base pairs were changed. Each of these unique pieces of DNA was then attached to a gene that produces a blue color change when activated. Finally, CRISPR was used to insert the mutated enhancers into the mouse genome to visualize the exact tissues and cell types in which they were active. Remarkably, nearly all (83%) of the enhancers could be mutated and still function normally.

Valentina Snetkova
“We were shocked to see how well these sequences still worked after introducing all these mutations,” said Valentina Snetkova, a postdoctoral researcher and first author of the study, which was published in Nature Genetics. “In one extreme case, we could change as many as 20% of all positions in the enhancer and it still showed some activity.”
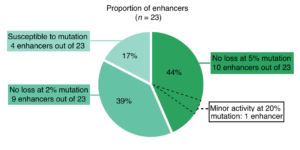
While the study dispels an explanation previously favored by many in the field, it also raises new questions. It is still possible that mutations in ultraconserved enhancers result in very subtle effects on gene expression that are difficult to detect, and yet decrease fitness. Alternatively, these sequences may have as-yet-unidentified functions, either regulatory or non-regulatory, that contribute to their extreme conservation.
“Even with this new data set in hand, we still don’t fully understand the driving force that caused these sequences to become ‘frozen’ in evolution. More work is needed to resolve this continued mystery,” said Visel, a senior scientist in EGSB.




