 Scientists gain important information about plant receptors from genomic analysis
Scientists gain important information about plant receptors from genomic analysis
Both plants and animals are targeted by rapidly evolving pathogens, including viruses, bacteria and fungi. Thanks to highly adaptive immune receptors, humans can mount a new antibody response towards infection or a vaccine over the course of a week. Plant immune receptors, however, do not typically change over the lifetime of an individual.
Lawrence Berkeley National Laboratory (Berkeley Lab) scientist Daniil Prigozhin collaborated with Ksenia Krasileva from University of California, Berkeley to study plant immune receptors using pan-genome sequencing, a technique which allows them to scan all genomes for every strain in a species within a particular branch on the tree of life. Their pan-genome analysis, published recently in The Plant Cell, showed that some plant immune receptors show a surprising degree of diversity within species. In addition, it allowed them to study how innate immunity evolves, where new receptor specificities come from, and the costs associated with making new receptors, such as the potential for autoimmunity. “I was not expecting to find such differences in diversity among these immune receptors,” said Prigozhin, research scientist in the Molecular Biophysics and Integrated Bioimaging (MBIB) Division. “All of the highly variable receptor subfamilies we studied have close relatives that are essentially unchanging within the species.”
Immune receptors can recognize either the physical surface of a pathogen-derived molecule (direct recognition) or the activity of a pathogen that leads to disease (indirect recognition). Indirect recognition provides a more evolutionarily robust form of immunity because it is easier for a pathogen to mutate a surface of an effector molecule than to change the way it causes disease. “The pan-genome immune receptor sequence analysis that we did can help us predict the mode of receptor function––that is, whether it is direct or indirect,” said Prigozhin.
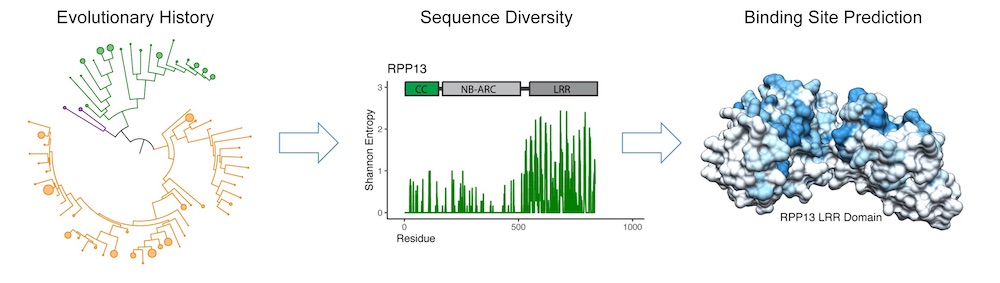
Pan-genome sequence data sheds light on the evolutionary history of plant NLR immune receptors (left) and helps identify receptor subfamilies of exceptional sequence diversity (center), allowing researchers to predict their target binding sites (right). (Daniil Prigozhin/Berkeley Lab)
Typically, when plants combine to form hybrids, the hybrids are healthier than the parents. Sometimes, the opposite is true and the resulting hybrids are less healthy than either parent. This is called hybrid incompatibility, which is a type of autoimmune reaction that often complicates breeding efforts. The pan-genome analysis of Prigozhin and Krasileva suggests which immune genes are likely to be responsible for negative interactions like these.
This was an important development, Prigozhin said, because prediction––from sequence data alone––of receptor function and behavior is an important step that can jump-start the analyses of immune systems of non-model plant species, including key crops. “These results will inform efforts to improve plant health by modifying the plants’ own immune receptors to achieve lasting protection against disease,” said Krasileva, assistant professor of plant and microbial biology at UC Berkeley. She added that this work is especially paramount for bioenergy crops where it might not be economical to use pesticides for pathogen control.
Environmental sustainability is indeed an important goal for the whole field of plant biology. Prigozhin elaborated, “In many areas around the globe, farmers cannot afford pesticide treatment and have to rely on resistant varieties that pathogens can defeat over time. In speeding up the derivation of new genetic resistance, scientists can be of help there, too.”




