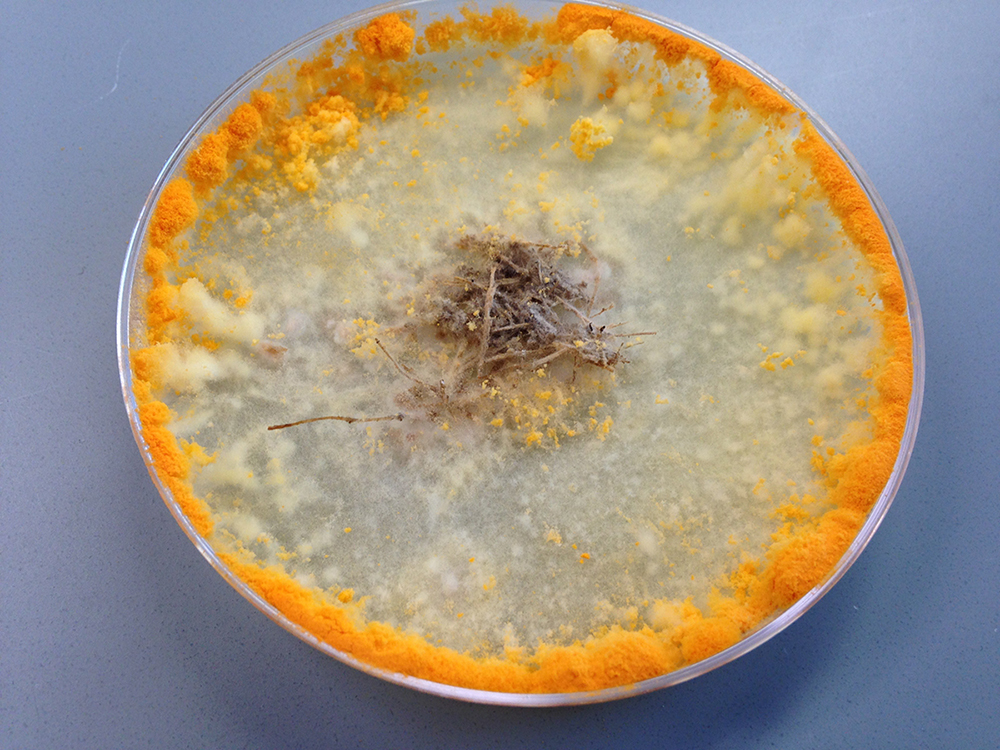
Filamentous fungi are like handymen who show up at a job site for a task that requires a flathead screwdriver with a full toolbox including Phillips and specialty screwdrivers, not to mention Allen wrenches. The fungi are similarly armed with a variety of PCWDEs to first break down the components of plant cell walls, which range from simple to complex carbohydrates, and then convert them into simple sugars. When faced with a veritable buffet of carbon sources, these fungi detect which complex chains are available; this information triggers pathways to determine which enzymes should be deployed in what order to most efficiently degrade the plant biomass.
In order to learn more about these regulatory networks in the model fungus Neurospora crassa, a team led by N. Louise Glass at the University of California, Berkeley and her postdoctoral fellow Vincent Wu worked with DOE Joint Genome Institute researchers. Glass is also the director of the Environmental Genomics & Systems Biology Division and her work was enabled in part through the Laboratory Directed Research and Development Program, as well as the JGI’s Community Science Program through a proposal akin to the Encyclopedia of DNA Elements (ENCODE) project, which is aimed at determining the activity, expression and regulation of protein-coding genes. This first paper integrating multi-omics data from this JGI fungal ENCODE proposal was recently published in the Proceedings of the National Academy of Sciences.
Read more here on the JGI website.