The unicellular green alga Chromochloris zofingiensis has the ability to shift metabolic modes from photoautotrophic (synthesizing food using light as energy source) to heterotrophic (obtaining food and energy from exogenous sources) in response to carbon source availability in the light. It also has the capacity—under certain conditions—to produce high amounts of commercially relevant bioproducts: notably, the ketocarotenoid astaxanthin, used in feed, cosmetics, and as a nutraceutical, and triacylglycerol (TAG) biofuel precursors.
Understanding how photosynthesis and metabolism are regulated in algae could, via bioengineering, enable scientists to reroute metabolism toward beneficial bioproducts for energy, food, and human health. To that end, Berkeley Lab Biosciences researchers used C. zofingiensis as a simple algal model system to investigate conserved eukaryotic sugar responses, as well as mechanisms of thylakoid breakdown and biogenesis in chloroplasts.
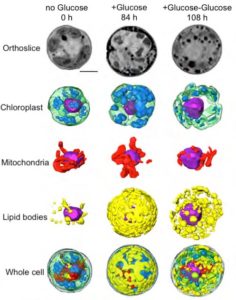
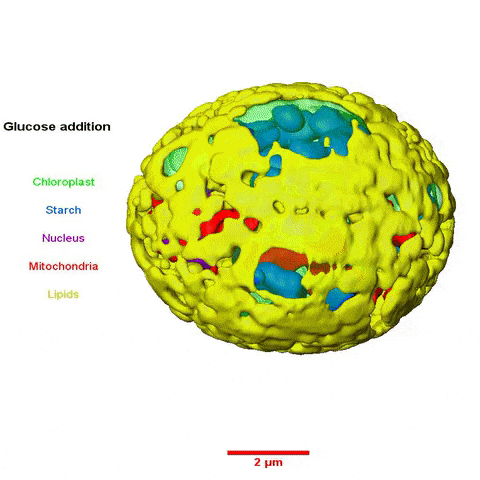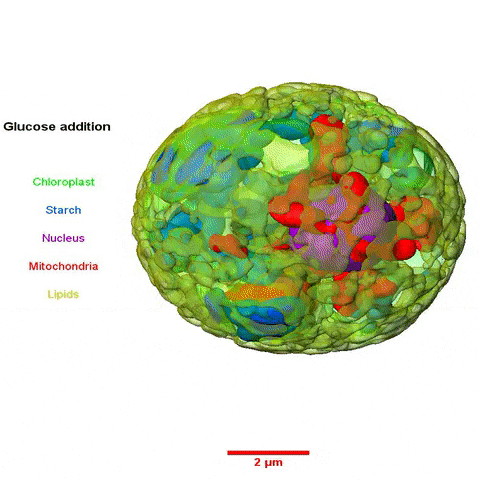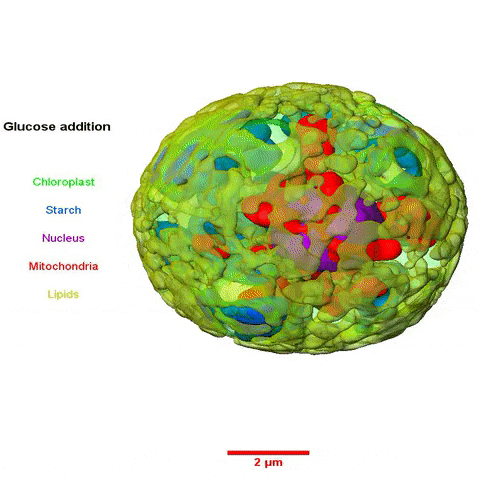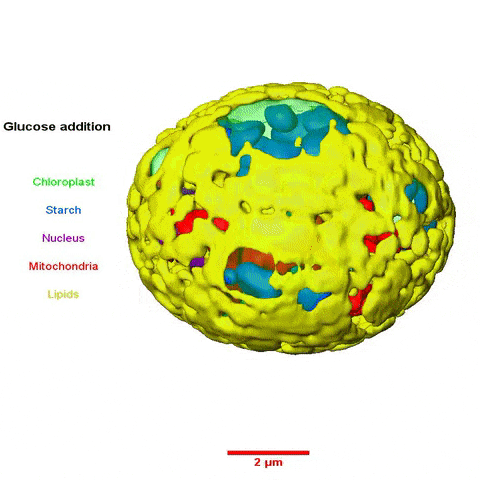
The collaboration among scientists from multiple Divisions was led by Krishna Niyogi, faculty scientist in Molecular Biophysics and Integrated Bioimaging (MBIB), and Melissa Roth, a former postdoctoral fellow in his lab who is now an assistant researcher at UC Berkeley. Using a variety of approaches—including cellular physiology analyses, transcriptomics, lipid analyses, transmission electron microscopy, soft X-ray tomography, and structured illumination microscopy—the team sought insight into what happens to C. zofingiensis with the addition of glucose while in the light.
In a paper published in the journal the Plant Cell they reported their observations of rapid and reversible repression of photosynthesis, dismantling of the photosynthetic apparatus, reduction in thylakoid volume, and increase in energy stores including lipids and starch. RNA sequencing revealed that changes in the transcriptome involving a third of all genes form the basis of this metabolic regulation.
The work made use of resources at the DOE Joint Genome Institute (JGI) for lipid analysis; the National Center for X-ray Tomography (NCXT) located at the Advanced Light Source (ALS) for cryo-soft X-ray tomography; and the DOE Joint BioEnergy Institute (JBEI) for electron microscopy analysis. Analysis of physiological experiments and RNA-Seq data was supported by the DOE Office of Science and sequencing was supported by a grant from the USDA National Institute of Food and Agriculture to first author Melissa Roth.
Other MBIB collaborators were: Carolyn Larabell, faculty scientist and director of NCXT; NCXT postdoctoral researchers Jian-Hua Chen and Andreas Walter; Manfred Auer, staff scientist and former director of physical analysis at JBEI; postdoctoral researcher Maria Mueller, and undergraduate students Nassim Ataii and Junha Song of the Auer lab; Masakazu Iwai, research scientist; and Daniel Westcott, graduate student in the Niyogi lab.
From Environmental Genomics and Systems Biology (EGSB), the collaborators were: Sabeeha Merchant, senior faculty scientist; Trent Northen, senior scientist and metabolomics technology group lead at JGI; as well as research scientist Benjamin Bowen and senior scientific associate Katherine Louie of the Northen group.