Genome-wide association (GWAS) and familial studies have found mutations that disrupt the function of KDM5 proteins in patients with autism spectrum disorders and related intellectual disabilities. Proteins belonging to this group regulate gene transcription by removing a methyl group from histone, a protein that associates with DNA in the nucleus and helps condense it into chromatin.
 Biological Systems and Engineering (BSE) Division researchers Jian-Hua Mao, Antoine Snijders, and Susan Celniker, in collaboration with a team of scientists led by Xingyin Liu at Nanjing Medical University (NMU) in China, used genetic tools to delineate how KDM5 contributes to autism and intellectual disability. Using the fruit fly (Drosophila melanogaster) as a model organism, they showed that reducing KDM5 activity causes intestinal barrier dysfunction and changes in social behavior that correlates with compositional changes in the gut microbiome. They further demonstrated that antibiotic treatment or feeding probiotic Lactobacillus species can partly rescue the social behavior, life span, and cell phenotype of KDM5-deficient flies.
Biological Systems and Engineering (BSE) Division researchers Jian-Hua Mao, Antoine Snijders, and Susan Celniker, in collaboration with a team of scientists led by Xingyin Liu at Nanjing Medical University (NMU) in China, used genetic tools to delineate how KDM5 contributes to autism and intellectual disability. Using the fruit fly (Drosophila melanogaster) as a model organism, they showed that reducing KDM5 activity causes intestinal barrier dysfunction and changes in social behavior that correlates with compositional changes in the gut microbiome. They further demonstrated that antibiotic treatment or feeding probiotic Lactobacillus species can partly rescue the social behavior, life span, and cell phenotype of KDM5-deficient flies.
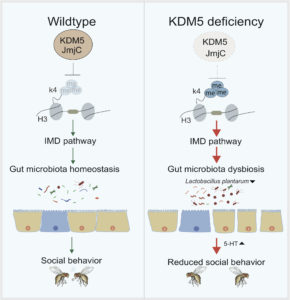
The study, published in the journal Cell Host Microbe, also revealed that KDM5 regulates innate immune signaling pathways and gut dysbiosis dependent on its histone demethylase function. This in turn affects the metabolism of the neurotransmitter serotonin (5-HT), impacting behavior through the gut-brain axis. The results of this study provide a new understanding of the pathological mechanism of autism spectrum disorders/intellectual disabilities from the perspective of multi-factor interaction, and suggest new possibilities for diagnosis and treatment.
Professor Liu Xingyin is corresponding author on the paper. The first authors are: graduate student Chen Kun, technician Xiaoting Luan, lecturer Qisha Liu, postdoctoral fellow Jianwei Wang, and technician Xinxia Chang NMU, and staff scientist Antoine Snijders of Berkeley Lab. Other contributors to the paper include: graduate student Zhou Dan of NMU; senior scientists Jian-Hua Mao and Susan Celniker of Berkeley Lab; Julie Secombe and Xiao Dong of Albert Einstein College of Medicine; and Zhibin Wang from Analysis and Testing Center of NMU.
This work was supported by NSFC grant 81671983 and 81871628, Natural science funding BK20161572 from Jiangsu province and starting package from NJMU (X.L.); Young scientist funding BK20161025 from Jiangsu province (Q.L. and J.W.); Lawrence Berkeley National Laboratory Directed Research and Development (LDRD) program funding under the Microbes to Biomes (M2B) initiative under contract DE AC02-05CH11231 (J.H.M., A.M.S. and S.E.C.); NIH grant R01GM112783 to (J.S.); and NSFC grant 31671311 (J.H.C.).




