Many DNA and RNA virus pathogens replicate in compartments in the cytoplasm of the cell that are made out of viral and cellular proteins that are called virus factories or viroplasms. Researchers on a team led by Mary Estes of the Baylor College of Medicine used rotavirus as a model to study some of the proteins involved in making these structures. Once the functions of proteins making the viroplasms are understood, this could lend insight into how other viruses make these factories and offer an avenue for disrupting virus production.
Banumathi Sankaran, a research scientist in the Berkeley Center for Structural Biology (BSCB) at the Advanced Light Source, was part of the team that published their findings in Proceedings of the National Academy of Sciences. Sankaran collected the X-ray data at the BCSB Beamline 5.0.1 that were used to solve the three-dimensional structures of nonstructural protein NSP2. Refinement of the structure model was carried out using programs within the Phenix software suite, developed under the guidance of Paul Adams, Director of Molecular Biophysics and Integrated Bioimaging (MBIB) Division.
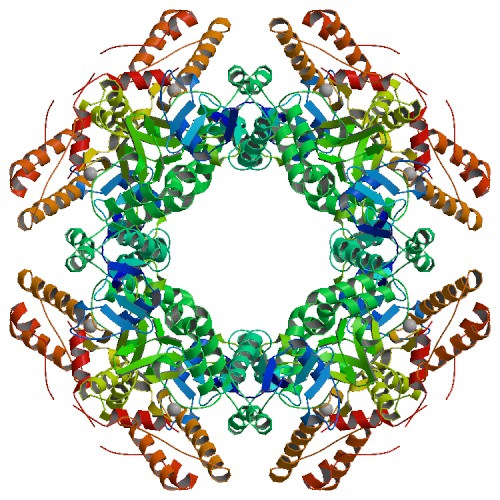
The atomic structure of the SA11 rotavirus NSP2 (PBD ID 6CY9), shown here, confirms the formation of a disulfide bridge in the wild-type protein.
Information from this structure helped elucidate the function of NSP2, which is able to add phosphorus to itself in a process called autophosphorylation and form an octamer that is a key component of the viroplasm. In addition, NSP2 can interact with other proteins and use phosphorylation to form additional building blocks. Because protein regulation by phosphorylation is a common biological mechanism, it is possible that similar phosphorylation-dependent virus factory assembly mechanisms exist for other viral pathogens.
X-ray data were collected at the BSCB through the Collaborative Crystallography program. This work was also supported by ALS-ENABLE, which is funded through the National Institute of General Medical Sciences. ALS-ENABLE integrates existing synchrotron structural biology resources to better serve researchers.