A team led by scientists at UC Davis that included Daniil Prigozhin, a research scientist in Molecular Biophysics and Integrated Bioimaging (MBIB), used artificial intelligence to help plants recognize a wider range of bacterial threats. The approach, reported in Nature Plants, may lead to new ways to protect important crops like tomatoes and potatoes from devastating diseases.
Like animals, plants have immune receptors that detect pathogens. One such receptor, called FLS2, detects the protein flagellin in the tails bacteria use to swim. But bacteria can change the amino acids in flagellin to evade detection
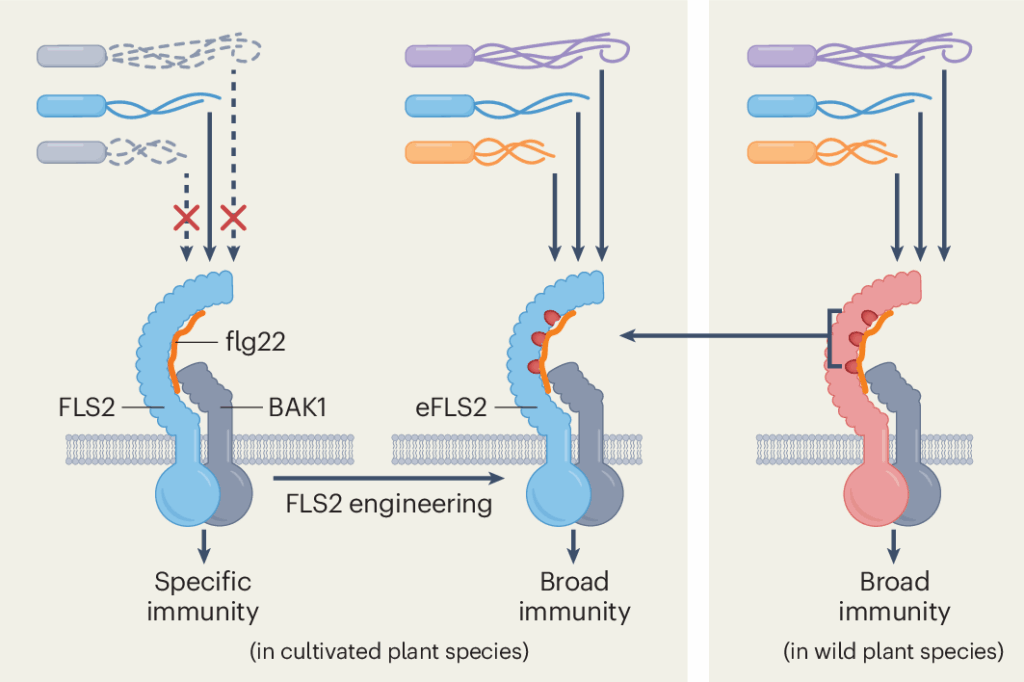
The researchers looked at versions of FLS2 found across plant lineages to identify ones that naturally recognize a broader range of bacteria. They then compared those to more narrowly focus versions to identify key amino acids that differed. Finally, they used AlphaFold—a tool developed to predict the 3D shape of proteins—to reengineer FLS2, essentially upgrading its immune system to catch more intruders.
“Evolutionarily ancient plant immune receptors that recognize conserved pathogen-associated molecules do change over time. As a result, variants found in different plant lineages have different specificity profiles that, in this case, could be identified with protein structure prediction,” said Prigozhin. “These natural innovations can be transferred to agriculturally important species and strengthen crop protection.”




