A team of researchers from the Joint BioEnergy Institute’s (JBEI) Feedstocks Division has, for the first time, developed a genome-scale way to map the regulatory role of transcription factors, the proteins that play a key role in gene expression and determining a plant’s physiological traits. Their work reveals unprecedented insights into gene regulatory networks and identifies a new library of DNA parts that can be used to optimize genetic engineering efforts in plants.
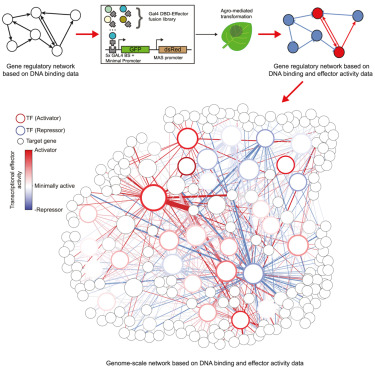
Study authors Niklas Hummel and Patrick Shih set out to develop a method to characterize a large number of transcription factors in a plant simultaneously. While methods to do this exist for other model organisms, such as animals, insects, and fungi, applying them to plants has been challenging, due to their complexity and the disrupting presence of cell walls.
To address this challenge, Hummel and Shih employed a transient expression system they had previously developed for building synthetic biology tools in plants. Here, they used the system to characterize, in parallel, a network of over 400 transcriptional effector domains in the tobacco plant Nicotiana benthamiana, a feat never before achieved in plant synthetic biology.




