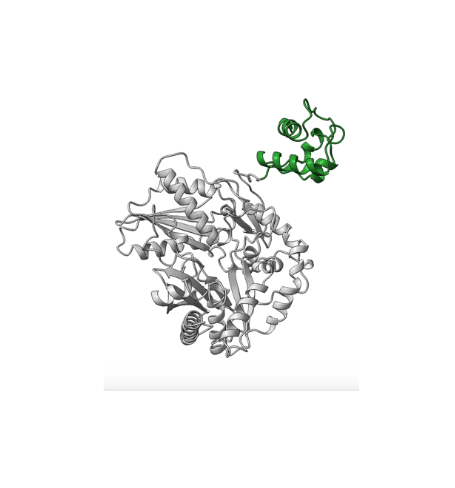
Many drugs used today are produced on enzymatic assembly lines—giant proteins known as nonribosomal peptide synthetases (NRPSs), which occur naturally in bacteria. Each “module” on the assembly line installs a single amino-acid building-block onto a growing peptide chain.
Today, NRPS molecular machines are used to produce important therapeutics such as penicillin, cyclosporine, and daptomycin (to name a few), and there is great interest in designing them to prepare next-generation drugs. Unfortunately, structural information on NRPSs is limited because of significant technical challenges.
In recent work, a team of researchers from the University of California, San Diego, collected multiple structures, or snapshots of the assembly line in action, using both cryogenic electron microscopy (cryo-EM) and protein crystallography, the latter done at the Berkeley Lab Advanced Light Source (ALS). The university team collaborated with the Berkeley Center for Structural Biology (BCSB) team on ALS Beamline 5.0.1 to define the structures, which revealed new details about how components of the assembly line work together at the molecular level.
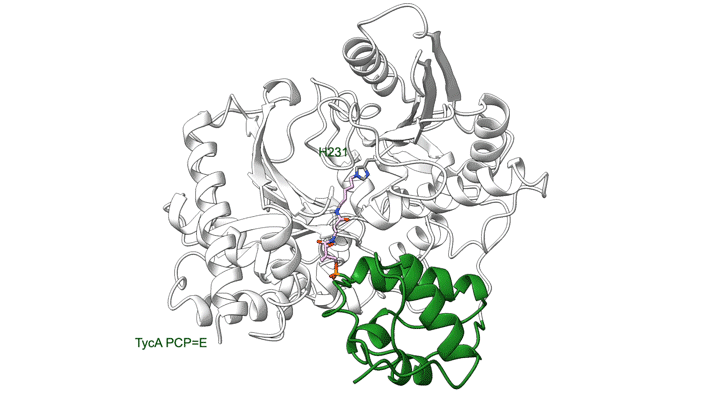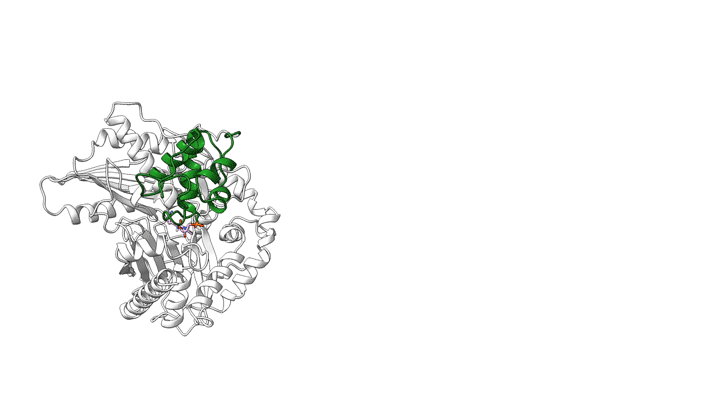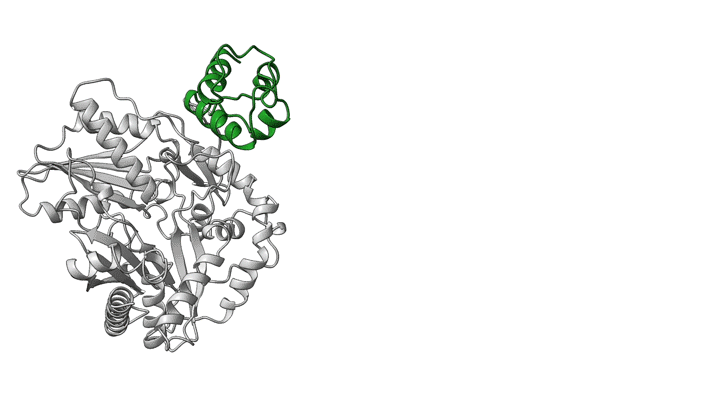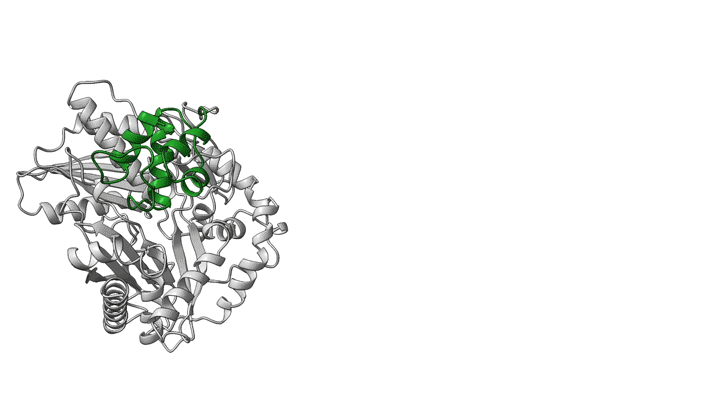
Overall, the structures demonstrate the mechanics between key modular components and the movement of a carrier protein from one step to the next. These results offer insights that could enable scientists to design and create new and improved medicines, such as antibiotics, using synthetic biology. The researchers hope this type of information will help with reengineering NRPSs to unlock, speed up, or increase the potency available in the massive diversity of medicinal peptides.
This article was adapted from an ALS Science Highlight.