HER2-positive breast cancer, which over-amplifies a protein called human epidermal growth factor receptor 2 (HER2), makes up about a third of all breast cancers. While HER2 helps control cell proliferation in normal cells, cancer cells with too much HER2 may multiply rapidly and spread more easily. HER2-positive breast cancer is not only aggressive, it is also frequently resistant to chemo-, radio-, and hormone therapies, with 70% of patients not responding to drug treatment. Moreover, a subset of patients who initially respond will ultimately develop resistance.
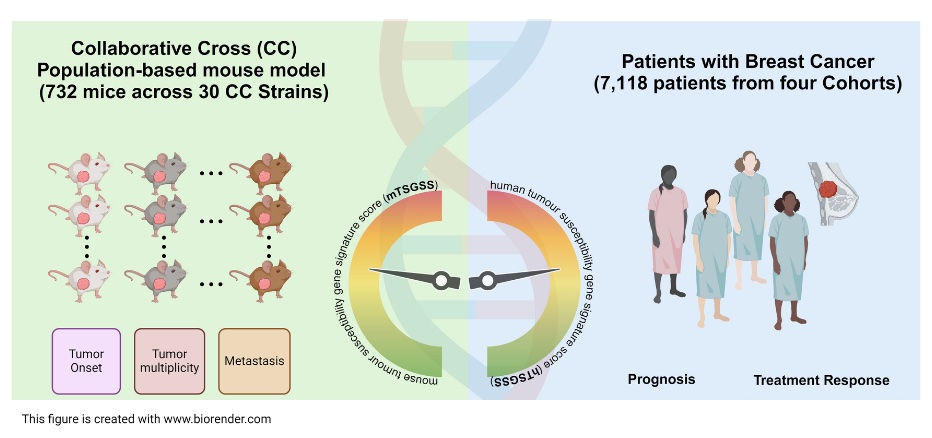
Scientists in Berkeley Lab’s Biological Systems and Engineering (BSE) Division were interested in understanding why some patients respond to treatment while others do not, with the ultimate goal of developing more tailored therapeutic strategies. In a previous study using mice, the team had determined that genetic background strongly impacts progression for this type of breast cancer. Such laboratory experiments with mice—which share about 97.5% of their functional DNA with homo sapiens—can be carefully controlled and monitored, providing a powerful means of parsing the contribution of genetics to cancer in people. But like many typical mouse studies, the researchers had only studied one strain of mouse, which means they likely missed important genetic variants at play in more diverse human populations. In their latest study, the researchers reexamined the effects of genetics on this form of breast cancer, this time using Collaborative Cross mice as a way to realistically approximate the genetic variety seen in human populations while still maintaining the precision of a lab-based experiment.
“Only a few genes had been studied and shown to be related to this cancer,“ said Jian-Hua Mao, a BSE senior scientist and lead author on the study. “We were interested in identifying additional genes that may be affecting tumor development and drug resistance.”
The researchers started by crossbreeding mice with the mouse version of the HER2+ gene with multiple strains of Collaborative Cross mice, resulting in a hybrid population of mice that had both the mutant breast cancer gene and a relatively high amount of genetic diversity. They then studied this heterogenous mouse cohort, tracking which mice developed tumors and how those tumors responded to chemotherapy. Analyzing the genomes of all the mice in the cohort revealed a set of 20 genes that were strongly associated with tumor onset, number and spread. The researchers combined these genes into a tumor-susceptibility gene signature that they hoped could be used as a scoring system to predict breast cancer prognosis and treatment outcomes in people.
To investigate whether the genes that they had identified in mice had clinical relevance for human patients, the team consulted population-scale genetic data made publicly available through The Cancer Genome Atlas (TCGA) project. They found that the human genes that functionally correspond to the 20 mouse genes they had identified all had altered expression patterns in breast cancer patients. Furthermore, expression patterns for those genes correlated with different clinical outcomes: Patients who scored low on the combined tumor-susceptibility metric had more favorable prognoses and were more responsive to chemotherapy. When tested on a large dataset of drug treatment responses in breast cancer patients, the team’s new signature outperformed available technology on optimizing drug selection. “It takes multiple steps between a fundamental discovery like ours and real clinical utility,” said Hang Chang, a BSE staff scientist and corresponding author on the study. “But that’s a very positive signal that lays the groundwork for future clinical evaluations and trials.”
In a clinical landscape where genetic technologies have improved and interest in personalized medicine have become more widespread, the researchers anticipate that their gene signature could be used as a biomarker for predicting patient treatment response. Anticipating how a given patient will respond to treatment may circumvent unnecessary drug exposures in patients unlikely to experience clinical advantages, and open up new avenues for treatments that are more patient-specific.
Both the scale of the study, with its mouse cohort of over 700, and its population-based approach were important design aspects that helped the team detect novel genes of interest and build toward an improved understanding of the genetic underpinnings of HER2+ breast cancer biology and drug resistance. Along with the new susceptibility genes identified in the study, the researchers observed several cases of cancer spreading to the lungs. While this progression is consistent with what clinicians commonly see for HER2+ breast cancer in humans, it had not been seen before in a mouse model. This finding suggests that the dynamics of cancer spreading to different organs may be mediated by genetic factors, and underscores the robustness of the Collaborative Cross approach for representing how this cancer behaves in genetically diverse human populations.
Additional BSE-affiliated contributors to this work include research scientist Jamie Inman, affiliate senior scientist Antoine Snijders, and affiliate scientist Pin Wang. The team also included collaborators from University of California, San Francisco and the University of Salamanca.




