As adults, we’ve likely all experienced at least one bout with the stomach flu. Known technically as gastroenteritis, this pesky ailment—which can sometimes turn deadly—is often caused by one of ten known rotaviruses (categorized A-J).
Rotaviruses in group A (RVA) are responsible for most gastrointestinal infections worldwide, particularly in children. Rotaviruses in group B (RVB) primarily infect adults and have been associated with large epidemic outbreaks and transmission to livestock. To date, RVAs are the best characterized, whereas little is known about the molecular mechanisms behind RVBs ability to recognize host cells and replicate.
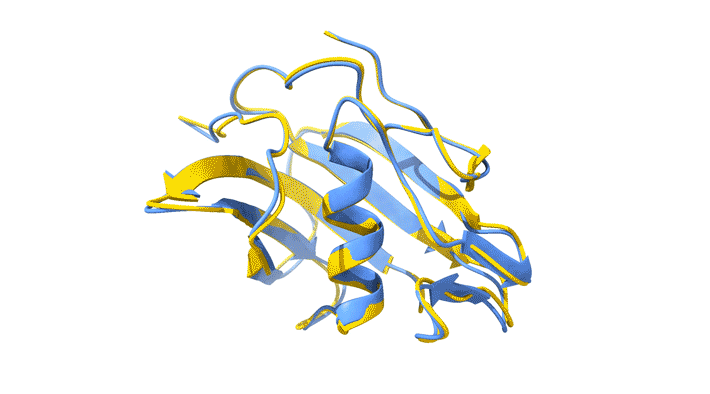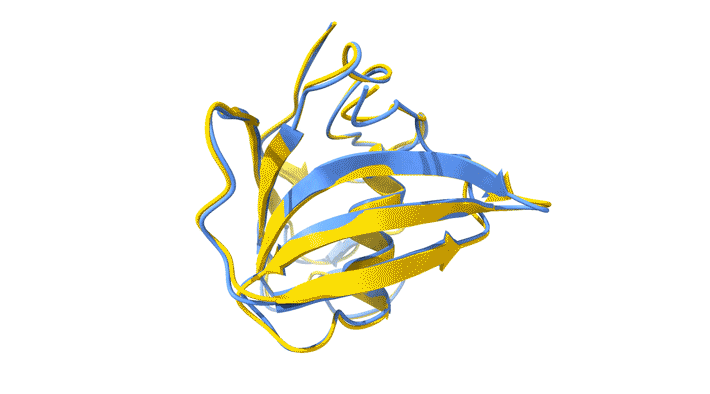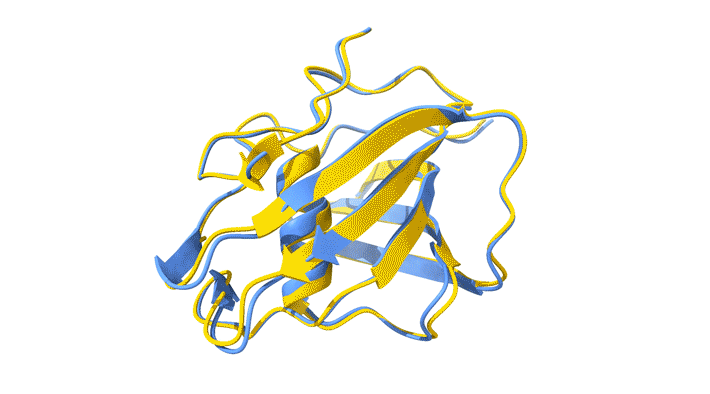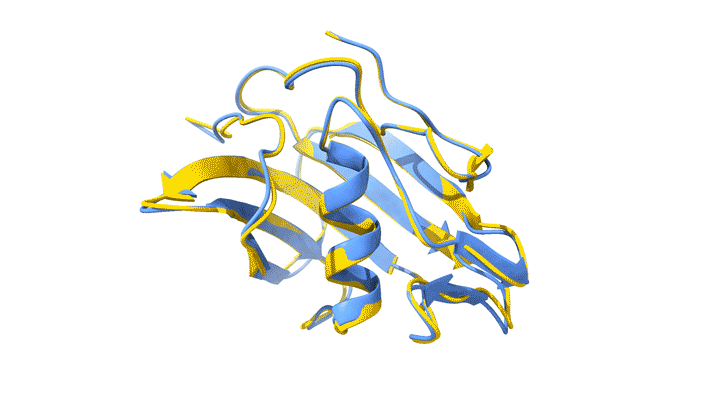
Using AlphaFold2, a deep-learning artificial-intelligence program for predicting protein structure, researchers from the Baylor College of Medicine discovered a new type of folding between molecules in the spike protein of RVBs. Banumathi Sankaran, a research scientist in the Molecular Biophysics and Bioimaging Division, validated these results by determining the structure at the Advanced Light Source (ALS) Beamline 5.0.1 in the Berkeley Center for Structural Biology (BCSB).
Sankaran works with the Baylor team through her Collaborative Crystallography program, which is part of ALS-ENABLE, an effort that integrates structural biology resources at the ALS. For this project, she screened samples and collected and processed the structural data to help validate the novel structure.
“I am quite excited about this work,” Sankaran said. “Getting good data is an important aspect of any structural work, and most of our efforts are directly related to us getting a better understanding of medical issues.”
Read more on this ALS Science Brief.