A recent study published in Nature Plants used a combination of genetic mutation and X-ray crystallography to reveal structural details of a key enzyme involved in plant signaling. The enzyme, a ubiquitin ligase named D3/MAX2, is responsible for transducing the signal from the plant hormone strigolactone, which regulates a plethora of processes in plant growth and development. D3/MAX2 can be thought of as a molecular switch, with open and closed states. By cycling between these two states, D3/MAX2 tags specific repressor proteins for degradation, thereby allowing strigolactone-response genes to be expressed at the correct time.
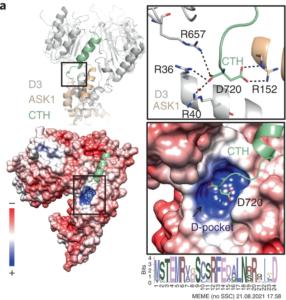
The X-ray crystallography was conducted at the Berkeley Center for Structural Biology (BCSB) and was partially funded by ALS-ENABLE. Using BCSB’s beamline 8.2.1. at the Advanced Light Source, followed by refinement and modeling using the Phenix software suite, the authors of the UC Davis study were able to solve the crystal structure of D3/MAX2. The structural data, coupled with experiments using a mutated D3/MAX2 locked in the open state, led the researchers to identify the specific part of the enzyme responsible for its activity in strigolactone signaling: a C-terminal helix that directly regulates the ubiquitin based degradation of repressor proteins.
Going a step further, the researchers also found that citrate, a primary plant metabolite, binds to D3/MAX2 and triggers the conformation shift at the C-terminal helix. This is the first time a primary metabolite has been shown to act as a direct regulator of this type of ubiquitin ligase and could point to an interesting avenue for further study.
Read the UC Davis press release and the ALS Science Highlight for more.




