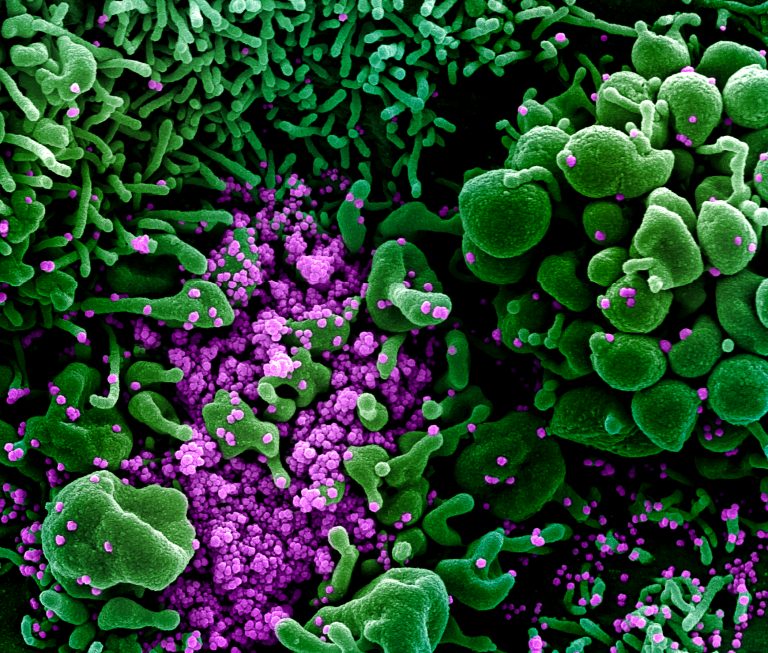
 As the SARS-CoV-2 pandemic continues and variants emerge, there is a need to increase the number of weapons in the arsenal to fight this virus. In addition, there is only one antiviral drug that has been approved by the Food and Drug Administration for emergency use in COVID-19, the infectious disease caused by SARS-CoV-2. To accelerate the pace of drug discovery, researchers from the Baylor College of Medicine employed previously constructed DNA-encoded chemistry technology (DEC-tec) libraries to identify several candidate molecules for further testing.
As the SARS-CoV-2 pandemic continues and variants emerge, there is a need to increase the number of weapons in the arsenal to fight this virus. In addition, there is only one antiviral drug that has been approved by the Food and Drug Administration for emergency use in COVID-19, the infectious disease caused by SARS-CoV-2. To accelerate the pace of drug discovery, researchers from the Baylor College of Medicine employed previously constructed DNA-encoded chemistry technology (DEC-tec) libraries to identify several candidate molecules for further testing.
In a paper published in the Proceedings of the National Academies of the Sciences USA, the team of scientists described how they screened their DEC-tec libraries containing four billion drug-like molecules to find small molecules and high affinity binders that would inhibit a protein of interest–in this case Mpro, the main protease of severe acute respiratory syndrome coronavirus 2, SARS-CoV-2. Mpro is a key enzyme responsible for preparing the functional proteins that act in propagation of the virus.
The rapid and efficient DEC-tec screening allowed researchers to identify a new inhibitor against the virus, CDD-1713. This molecule was further tested to determine its affinity for Mpro and to ensure it would not bind to four common classes of human proteases.
After clearing these early hurdles, the structure of Mpro in complex with CDD-1713 was solved using X-ray crystallography. Study co-author Banumathi Sankaran, a research scientist in the Molecular Biophysics and Bioimaging Division, used the Advanced Light Source (ALS) beamlines in the Berkeley Center for Structural Biology (BCSB) to collect the crystallography data. These data allowed the researchers to determine that CDD-1713 inhibits the activity of Mpro by binding in the active site of this enzyme.
The researchers tested the ability of CDD-1713 and a related molecule to prevent the death of cells exposed to SARS-CoV-2 using a real-time cell analysis assay. The specific interactions that were measured in the crystal structure of the complex will help with further refinement to improve the inhibitory activity of this molecule.
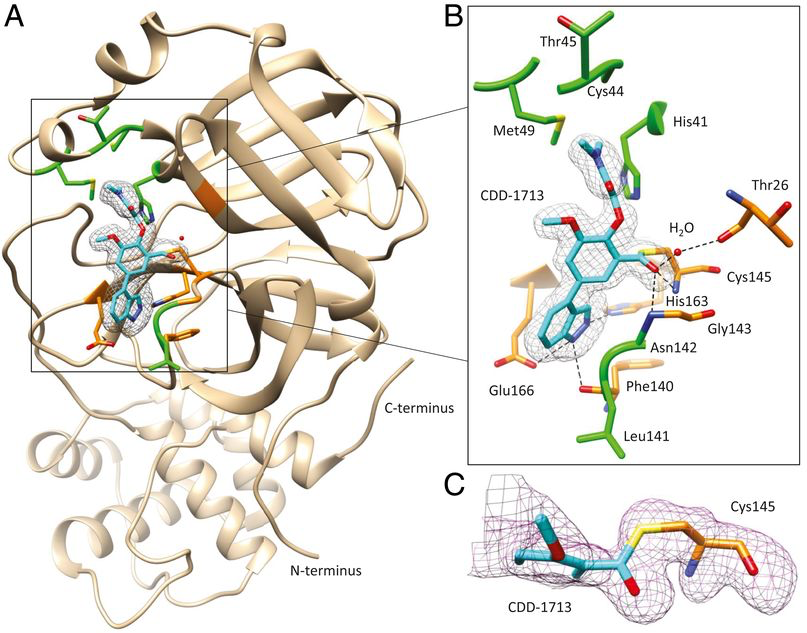
(A) This is the crystal structure of Mpro (tan) in complex with CDD-1713 (cyan). The active site of the protease is located within the square. (B) In the magnified view of the catalytic center, carbon atoms of the inhibitor are cyan, nitrogen atoms are blue, and oxygen atoms are red. The Mpro amino acid residues involved in CDD-1713 binding are shown as stick models and labeled. (C) The difference Fourier map shows the relative position of CDD-1713 in the active site of Mpro. (Credit: Adapted from Chamakuri, Lu, Ucisik, et al., PNAS, 2021.)
This work, which rapidly identified a new class of inhibitors to Mpro, was led by Martin Matzuk, Timothy Palzkill, and Damian Young at Baylor College of Medicine. Sankaran leads the Collaborative Crystallography mail-in service at the ALS, which is run under the aegis of the ALS-ENABLE program. Refinement of the structure model was carried out using programs within the Phenix software suite, developed under the guidance of Paul Adams, Associate Laboratory Director for Biosciences.