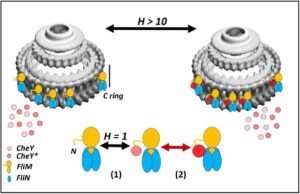
 The protein CheY plays a role in relaying sensory signals from chemoreceptors to the rotary motor at the base of the tail-like appendage, or flagellum, that protrudes from the cell body of certain bacteria and eukaryotic cells. It has been studied as a model for dissecting the mechanism of allostery—the process by which the binding of biological macromolecules (mainly proteins) at one location regulates activity at another, often distant, functional site. When it is transiently phosphorylated in response to chemotactic cues, CheY’s binding affinity for a flagellar motor switch protein called FliM is enhanced. CheY binding to FliM changes the direction of flagellar rotation from counterclockwise to clockwise.
The protein CheY plays a role in relaying sensory signals from chemoreceptors to the rotary motor at the base of the tail-like appendage, or flagellum, that protrudes from the cell body of certain bacteria and eukaryotic cells. It has been studied as a model for dissecting the mechanism of allostery—the process by which the binding of biological macromolecules (mainly proteins) at one location regulates activity at another, often distant, functional site. When it is transiently phosphorylated in response to chemotactic cues, CheY’s binding affinity for a flagellar motor switch protein called FliM is enhanced. CheY binding to FliM changes the direction of flagellar rotation from counterclockwise to clockwise.
Using X-ray footprinting with mass spectroscopy (XFMS), a team led by Shahid Khan, a senior scientist with the Molecular Biology Consortium, established that CheY changes shape when it tethers to the motor, and further parsed the contribution of phosphorylation to this shape change. The results of the XFMS experiments validated atomistic molecular dynamics (MD) predictions of the architecture of the allosteric communication network, marking the first time that XFMS has been used to validate protein dynamics simulations at single-residue resolution sampled over the complete protein.
“Because transient phosphorylation in the CheY superfamily determines motile responses of photosynthetic and nitrogen-fixing bacteria to light and nutrient gradients, respectively, detailed knowledge of allosteric communication within this family can be used to engineer strains with optimized dispersal strategies to increase the efficiency by which energy is harvested by these microbes,” said Khan.
Several individuals in the Biosciences Area contributed to the XFMS experiments, which took place at the Advanced Light Source (ALS) beamline 3.2.1 and the Joint BioEnergy Institute (JBEI). They are: Sayan Gupta, a research scientist in the Molecular Biophysics and Integrated Bioimaging (MBIB) Division; Corie Ralston, a staff scientist in MBIB and Facility Director, Biological Nanostructures, at the Molecular Foundry; Yan Chen, a project scientist in the Biological Systems and Engineering Division (BSE); and Chris Petzold, a staff scientist in BSE and Director of Functional Genomics and Deputy VP of Technology at JBEI.
In addition, collaborators at the University of Utah prepared and characterized CheY-FliM fusion proteins used to probe the attachment of the FliM N-terminal peptide tether to CheY. In the absence of phosphorylation, this attachment is weak and could not have been evaluated to study the specific effect of phosphorylation were the peptide not fused to the CheY protein.
Read the paper in Biophysical Journal.




