In early January, the Chinese scientists who sequenced the viral SARS-CoV-2 genome made that data public. Now researchers are scrutinizing this data for insight into the COVID-19 disease, using supercomputing resources to meet the challenge.
For example, City of Hope’s Beckman Research Institute, which is already involved with a number of SARS-CoV-2 genome analyses and related projects, and City of Hope affiliate TGen (Translational Genomics Research Institute) are using supercomputing resources at Berkeley Lab’s National Energy Research Scientific Computing Center (NERSC) to aid in these efforts.
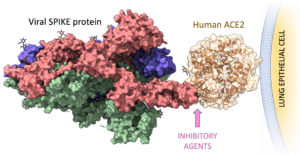
Crystal structure of SARS-Cov-2 SPIKE protein in complex with human ACE2 on the surface of the lung epithelial cells. The binding interface is the target for the computational design of inhibitory agents to prevent viral infection. (Credit: Supriyo Bhattacharya/Beckman Research Institute at City of Hope)
The City of Hope research targets the entire viral genome, with particular scrutiny on key viral proteins, initially focused on the Spike and M protein, as potential drug targets with the hope of developing antiviral treatments. Biological Systems and Engineering (BSE) Division Director Blake Simmons is involved in facilitating the project, maintaining regular contact with the City of Hope researchers, and project planning; he will also assist in the analysis of the results.
“The collaboration with City of Hope will use the computing resources of NERSC to identify small molecules that can be used to assist in the diagnosis of COVID-19, design peptide libraries to mimic virus-host interactions, and annotate the virulence of the Italian strain as compared to the Chinese strain of COVID-19,” said Simmons.




