Microbiomes play critical roles in ecosystem and human health, but direct characterization studies of microbial communities in their native states are limited by high cost, limited replicability, and the inability to use reductionistic technologies, such as engineered microbes. Most of what is currently known about the function of microbial genes has come from studying microbes in isolation. And yet, microbial communities—whether they reside in the human gut, terrestrial soils, or ocean sediment—are highly heterogeneous and dynamic, exhibiting variability in composition and function across space and over time.
Science grapples with the complexity of the natural world using models: simplified proxy systems that aim to make particular features tractable to controlled laboratory studies. Many of the fundamental principles of biology have been discovered thanks to the development of model organisms—such as the fruitfly Drosophila melanogaster; the nematode Caenorhabitis elegans; the zebrafish Danio rerio; the plants Zea mays (corn) and Arabidopsis thaliana (cress); and the microorganisms Escherichia coli (a bacterium) and Saccharomyces cerevisiae (yeast). However, the establishment of these and other model organisms in wide use today was a lengthy process that spanned several decades and, in many cases, involved multiple generations of scientists.
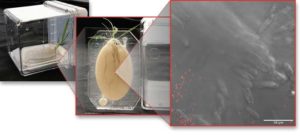
In EcoFABs, plants are grown with a tailored root microbiome. Here, Brachypodium distachyon with fluorescent Pseudomonas simiae growing next to the plant’s root hairs. (Credit: T. Northen, D. Chiniquy, L. Jabusch, Berkeley Lab/JGI; E. Dewalt/Springer Nature)
To date, standardized and reproducible model systems are largely nonexistent in most areas of microbiome science. For this reason, asserts Berkeley Lab Biosciences’ Trent Northen, “there is an urgent need for the microbiome community to develop a few widely accepted and reproducible model microbial ecosystems.”
Northen is the lead author on a paper published in Nature Methods that outlines a vision for fabricated model microbial ecosystems (EcoFABs) and their potential impact on microbiome science. The paper was co-authored with collaborators from more than a dozen institutions, including other national labs, universities, and the National Institute of Standards and Technology (NIST). Ben Brown, Environmental Genomics and Systems Biology (EGSB) co-deputy for science, also was among the authors.
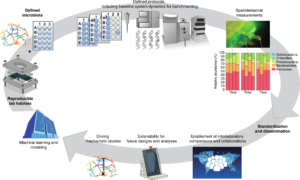
Vision for EcoFABs and their impact on microbiome science. (Courtesy Nature Methods)
EcoFABs are composed of defined communities of microbiota, laboratory habitats, and protocols for cultivation and spatiotemporal analysis. Last spring, Northen’s groups in EGSB and at the DOE Joint Genome Institute (JGI) published detailed video protocols for EcoFABs. Northen posits that “standardized EcoFABs will enable researchers to build on each other’s research and perform mechanistic studies.” He and his colleagues have formed a steering committee to oversee and expedite these important activities.
“EcoFAB datasets will be a boon for computational ecology,” adds Brown. “We have rarely had sufficiently large, homogeneous datasets to leverage advanced statistical machine learning algorithms. EcoFABs will change that.”
Studying engineered microbiomes in ecologically relevant community contexts will provide mechanistic insights into genes, pathways, and metabolites. And this improved understanding may ultimately help predict and harness plant-microbe interactions for applications in agriculture and sustainable bioenergy.
Northen is also quoted in an accompanying Technology Feature appearing in the same issue of the journal.




