Researchers led by Diane Dickel have successfully adapted an open-source RNA analysis platform to study gene expression in individual plant cells. The method, called Drop-seq, was developed at Harvard Medical School in 2015 and had previously been used only in animal cells.
Researchers led by Diane Dickel have successfully adapted an open-source RNA analysis platform to study gene expression in individual plant cells. The method, called Drop-seq, was developed at Harvard Medical School in 2015 and had previously been used only in animal cells.
Dickel, who studies mammalian genomics in the Environmental Genomics and Systems Biology (EGSB) Division, has been using Drop-seq on animal cells for several years. She was impressed by the platform’s ease of use and efficacy, and became intrigued by the possibility of using it on plant cells. For many of the genes in plants, we have little to no understanding of what they actually do, she explained. “But by knowing exactly what cell type or developmental stage a specific gene is expressed in, we can start getting a toehold into its function.”
Unlike animal cells, plant cells have tough walls that make it harder to open them up for genetic study. To run plant cells through a single-cell RNA-seq analysis, their walls must be broken down via a process called protoplasting. This is no easy feat because cells from different species—and even different parts of the same plant—require unique enzyme cocktails. Some plant biologists even expressed skepticism that plant cells would be altered too significantly by the protoplasting process to provide meaningful insight into their normal function.
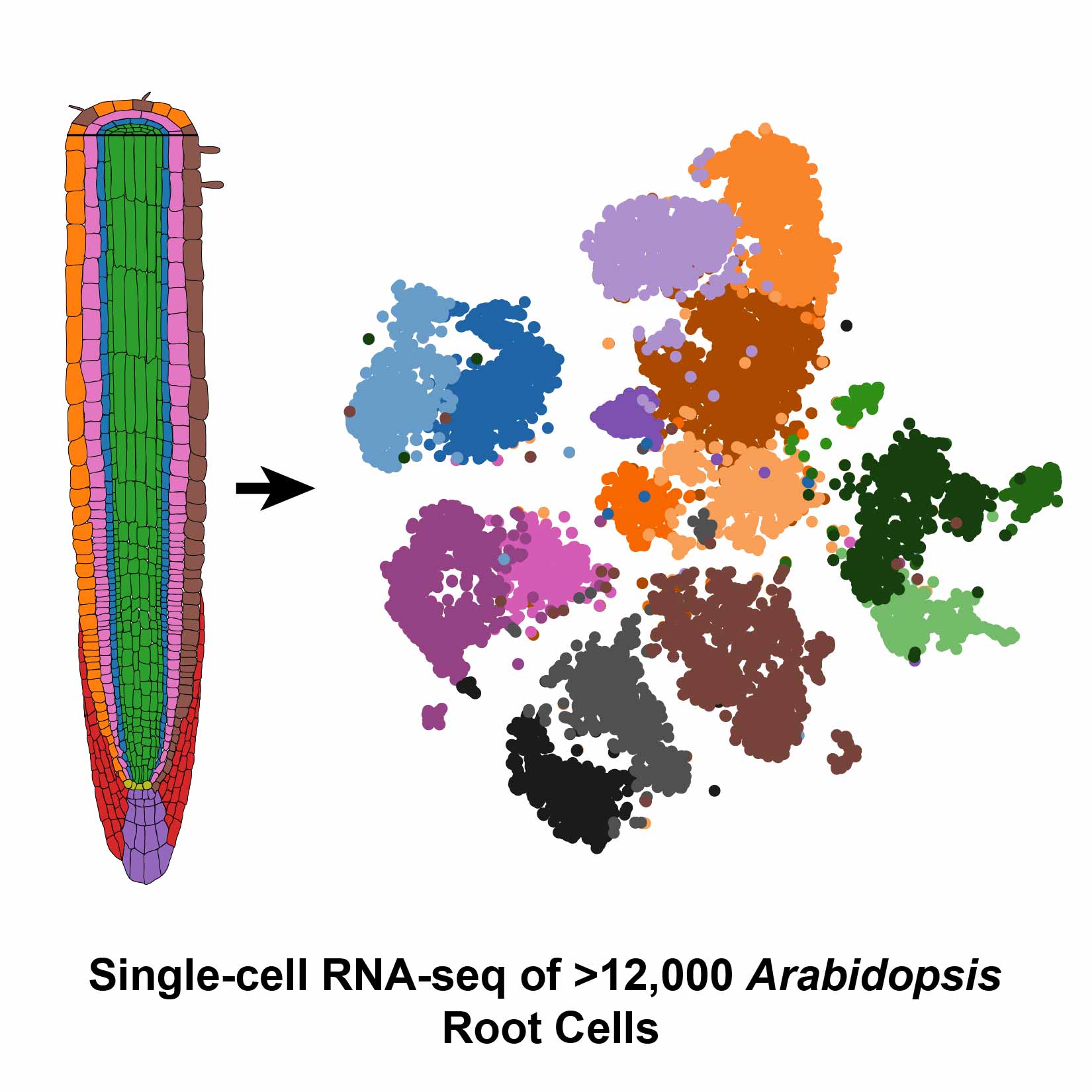
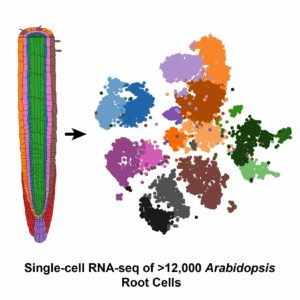 Undeterred by these challenges, Dickel and her colleagues at the DOE Joint Genome Institute (JGI) teamed up with researchers from UC Davis who had perfected a protoplasting technique for root tissue from Arabidopsis thaliana (mouse-ear cress). After preparing samples of more than 12,000 Arabidopsis root cells, the group was thrilled when the Drop-seq process went smoother than expected. Their results were published in Cell Reports.
Undeterred by these challenges, Dickel and her colleagues at the DOE Joint Genome Institute (JGI) teamed up with researchers from UC Davis who had perfected a protoplasting technique for root tissue from Arabidopsis thaliana (mouse-ear cress). After preparing samples of more than 12,000 Arabidopsis root cells, the group was thrilled when the Drop-seq process went smoother than expected. Their results were published in Cell Reports.
“Part of Berkeley Lab’s mission is to better understand how plants respond to changing environmental conditions, and how we can apply this understanding to best utilize plants for bioenergy,” noted first author Christine Shulse, who is currently a JGI affiliate. “In this work, we generated a map of gene expression in individual cell types from one plant species under two environmental conditions, which is an important first step.”
This research was funded by the Laboratory Directed Research and Development (LDRD) program. The other Biosciences Area authors were: Doina Ciobanu (JGI), Benjamin Cole (JGI), Junyan Lin (JGI), Ronan O’Malley (EGSB/JGI), Yuko Yoshinaga (JGI), and Yiwen Zhu (EGSB).
Read more in the Berkeley Lab News Center.