 UC Berkeley and Berkeley Lab scientists have expanded the CRISPR gene-editing toolkit with the addition of a new, compact CRISPR-associated (Cas) protein—the RNA-guided “scissors” that snip DNA—and a modification of the Cas9 protein to give it an “on” switch for better control.
UC Berkeley and Berkeley Lab scientists have expanded the CRISPR gene-editing toolkit with the addition of a new, compact CRISPR-associated (Cas) protein—the RNA-guided “scissors” that snip DNA—and a modification of the Cas9 protein to give it an “on” switch for better control.
In December 2016, a team led by UC Berkeley professors and Berkeley Lab faculty scientists Jennifer Doudna and Jill Banfield reported the discovery of two novel CRISPR-associated (Cas) proteins identified through metagenomic analysis of microbial DNA from groundwater samples. They were given the placeholder names CasX and CasY, pending further analysis and classification.
Now, a group of researchers led by Doudna, a faculty scientist in Molecular Biophysics and Integrated Bioimaging (MBIB), and Eva Nogales, a senior faculty scientist in MBIB, has determined the unique structure of CasX and figured out how it works. They recently reported their findings in the journal Nature.
Like Cas9, the original and most commonly used CRISPR effector, CasX is capable of inducing RNA-guided, site-specific double strand breaks in DNA. However, CasX is about 40 percent smaller than Cas9. The small but mighty enzyme was found to be an effective genome editor in both E. coli and human cells, establishing it as a third, distinct CRISPR-Cas platform.
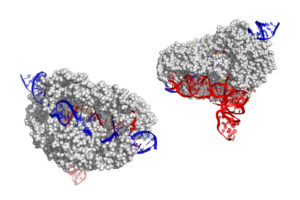
Co–first authors Jun-Jie Liu, a former joint postdoc in the labs of Doudna and Nogales, and Natalia Orlova, a former postdoc in Doudna’s lab, captured hundreds of thousands of images of CasX using cryo-electron microscopy (cryo-EM). The resulting atomic models revealed an unexpected architecture and organization dominated by the RNA scaffold.
Read more about CasX in the UC Berkeley press release.
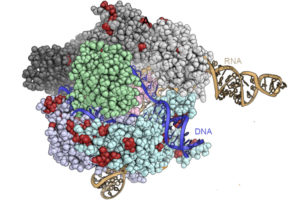 In a separate study, Doudna’s group, in collaboration with UC Berkeley Molecular and Cell Biology colleague David Savage’s lab, modified the seminal Cas9 enzyme with a molecular “lock,” a length of protein that must be snipped before it can bind and cut DNA. The redesigned enzyme, which the researchers refer to as ProCas9, is activated by a specific protease “key” that recognizes that protein chain.
In a separate study, Doudna’s group, in collaboration with UC Berkeley Molecular and Cell Biology colleague David Savage’s lab, modified the seminal Cas9 enzyme with a molecular “lock,” a length of protein that must be snipped before it can bind and cut DNA. The redesigned enzyme, which the researchers refer to as ProCas9, is activated by a specific protease “key” that recognizes that protein chain.
According to their paper published in the journal Cell, ProCas9 can be used to target specific cells during CRISPR gene editing. The researchers tested this system in living cells, and found that they could successfully lock ProCas9 and release its action using protein keys from viruses that threaten human health (Zika, West Nile), as well as viruses that cause major loss of food crops.
Adam Arkin, a senior faculty scientist in Environmental Genomics and Systems Biology (EGSB) and Dean A. Richard Newton Memorial Professor of Bioengineering at UC Berkeley was also an author on the paper.
Read more about ProCas9 in the UC Berkeley press release.