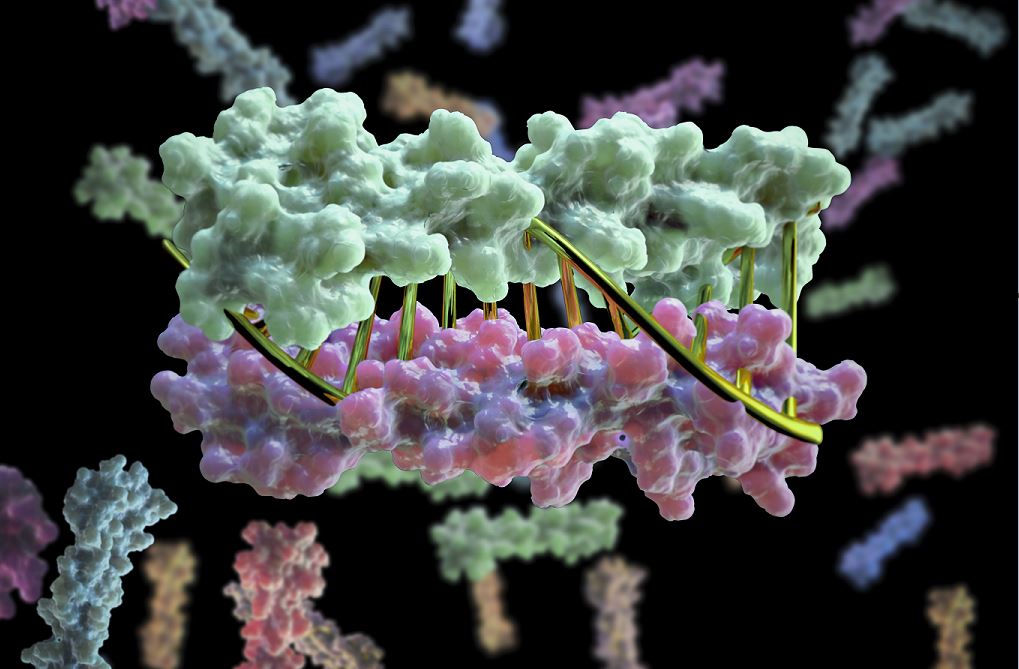
Bioscientists at the Advanced Light Source (ALS) at Berkeley Lab lent their expertise to a project led by scientists at the University of Washington to design proteins in the lab that zip together like DNA. The technique could enable the design of protein nanomachines to help diagnose and treat disease, allow for more precise engineering of cells, and perform a variety of other tasks.
The research, which was published online in the journal Nature, was performed at UW Medicine’s Institute of Protein Design, directed by David Baker. The team developed new protein design algorithms that produce complementary proteins that precisely pair with each other using the same chemical language that allows DNA strands to come together and form hydrogen bonds to create the double helix.
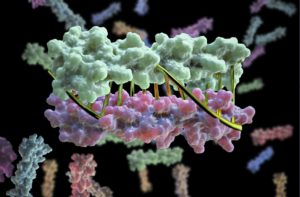
Scientists have developed a technique for designing proteins in the lab that provides a precise, programmable way to control how biomolecular machines interact. (Credit: UW Medicine Institute for Protein Design)
UW researchers worked with X-ray crystallography datasets that were collected at the ALS on Berkeley Center for Structural Biology (BCSB) beamlines 8.2.1 and 8.2.2. to determine the atomic structure of select complementary proteins. Models were generated and refined using programs within the Phenix software suite, developed under the guidance of Paul Adams, Division Director of Molecular Biophysics and Integrated Bioimaging (MBIB). Small-angle X-ray scattering (SAXS) datasets were obtained on the Structurally Integrated Biology for Life Sciences (SIBYLS) beamline 12.3.1. The UW team made use of the unique high-throughput SAXS capabilities of the SIBYLS beamline to verify their macromolecular engineering goals.
In the Nature paper reporting their findings, the UW team thanked the following MBIB staff: Banumathi Sankaran, research scientist at BCSB; and SIBYLS group members Kathryn Burnett (research assistant), Scott Classen (research scientist), Michal Hammel (research scientist), Greg Hura (research scientist, ALS-ENABLE SAXS core lead), Tad Ogorzalek (senior scientific engineering associate), John Tainer (visiting faculty, group leader), and Jane Tanamachi (program manager).
X-ray data were collected at the BSCB through the Collaborative Crystallography program. DOE’s Office of Biological and Environmental Research supports SIBYLS as part of the Integrated Diffraction Analysis Technologies program. ALS-ENABLE, which is funded through the National Institute of General Medical Sciences, integrates existing synchrotron structural biology resources to better serve researchers.
Read the UW press release.