Bacterial microcompartments (BMCs) are organelles that encapsulate portions of metabolic pathways, like miniature factories. They’re found across diverse phyla and do different things depending on the host. Scientists want to retrofit these factories to perform desired functions, such as producing biofuels, industrial materials, or nanoscale medical devices. But current technologies to manipulate BMCs, which consist of an enzymatic core surrounded by a shell made up of protein tiles, have limitations.
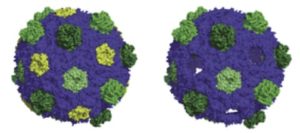
Left: BMC shells are like soccer balls, made of different types of protein tiles. Right: a BMC missing its yellow tiles. (Credit: Andrew Hagen)
In a recently published Nature Communications paper, researchers affiliated with Berkeley Lab’s Environmental Genomics and Systems Biology (EGSB) Division and Michigan State’s MSU-DOE Plant Research Laboratory present two new methods they’ve developed to facilitate the construction of synthetic versions.
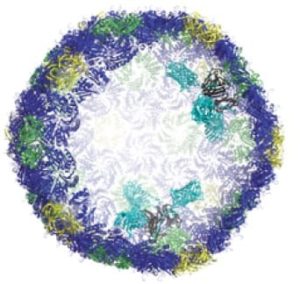
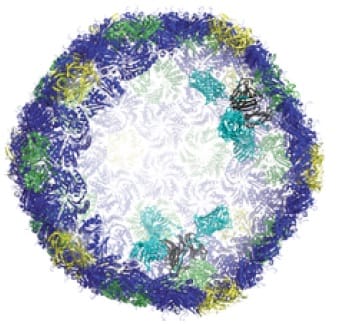
BMC cutout: in black, docking sites inside the wall; in turquoise, attached machinery. (Credit: Andrew Hagen)
The first, complementation-based affinity purification (CAP), quickly screens for the assembly and extraction of the shells. And the second, encapsulation via covalent-linkage (EnCo), helps to predictably insert custom enzymes. Using the two methods in concert, the team led by faculty scientist Cheryl Kerfeld has produced a factory shell, inserted machinery, sealed it off, and extracted it in experiments. Postdoctoral fellow Andrew Hagen, research associate Nancy Sloan, and senior research associate Markus Sutter also contributed to the research.
Read more from the MSU-DOE Plant Research Laboratory.