The US Department of Energy Systems Biology Knowledgebase, or KBase, is a collaborative, open software and data platform designed to meet the grand challenge of predicting and designing biological function from the biomolecular (small scale) to the ecological (large scale). The collaboration is led by Berkeley Lab and includes participation from Argonne (ANL), Brookhaven, and Oak Ridge National Labs (ORNL) along with Cold Spring Harbor Laboratory. Adam Arkin, a senior faculty scientist in Biosciences’ Environmental Genomics & Systems Biology Division, is the lead principal investigator; Robert Cottingham at ORNL and Christopher Henry at ANL are co-principal investigators.
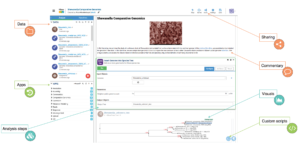
In KBase, anyone can upload and analyze their data and share their results and commentary with other researchers, who can rerun or build upon their work, accelerating the pace of scientific progress.
In a letter to the editor published July 6 in Nature Biotechnology, the KBase team presented a comprehensive overview of the platform and an assessment of its scientific impact. The paper describes the unique features and infrastructure of the platform, in addition to highlighting scientific use cases that demonstrate its significance for biology research. A series of linked scenarios, wherein two scientists use KBase to perform collaborative systems biology analysis, illustrate how it can help researchers accomplish more together than they could individually, with less work and in less time.
In addition to the collaborations with various scientific focus areas, national laboratories, and bioenergy research centers, key KBase collaborators specifically at Berkeley Lab include researchers affiliated with the DOE Joint Genome Institute, as well as NERSC.
Read more in this KBase Research Highlight.




