While advances in sequencing technologies have enabled researchers to access the genomes of thousands of microbes and make them publicly available, the task of assigning functions to the genes uncovered has lagged behind due to the limited capacity of functional analysis approaches.
To help overcome this bottleneck, Berkeley Lab researchers, led by Adam Arkin and Adam Deutschbauer in Biosciences’ Environmental Genomics and Systems Biology (EGSB) Division and Matthew Blow at the DOE Joint Genome Institute (JGI), have developed a workflow that enables large-scale, genome-wide assays of gene importance across many conditions.
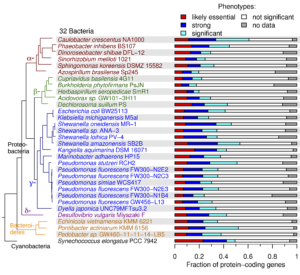 The team tested the workflow—which combines high-throughput genetics and comparative genomics—on 32 bacteria from various genera to identify mutant phenotypes for thousands of genes with previously unknown functions. To more efficiently generate mutant libraries for each bacterium, they refined a DNA bar-code sequencing approach known as RB-TnSeq (randomly bar-coded transposon sequencing).
The team tested the workflow—which combines high-throughput genetics and comparative genomics—on 32 bacteria from various genera to identify mutant phenotypes for thousands of genes with previously unknown functions. To more efficiently generate mutant libraries for each bacterium, they refined a DNA bar-code sequencing approach known as RB-TnSeq (randomly bar-coded transposon sequencing).
The results of what was, by far, the largest functional genomics study of bacteria to date, were published in the journal Nature.
Read more in the Berkeley Lab News Center.




