Though once thought to be “junk DNA,” enhancers are extremely important, regulating the expression of specific genes that ultimately determine a cell’s properties and functions. However, there are many more enhancers than genes and their relationship is unclear. A team of scientists in the Environmental Genomics & Systems Biology Division have turned their attention to this relationship and the overall importance of enhancers to development. By better linking the genomic complement of an organism with its expressed characteristics, their work offers new insights that further the growing field of systems biology, which seeks to gain a predictive understanding of living systems.
In two recent studies, one published in Cell on January 18 and the other reported on January 31 in Nature, the researchers answered a long-standing question about the role of enhancers. Diane Dickel, research scientist, co-led the Cell studies with Len Pennacchio and Axel Visel, both senior scientists, investigated enhancers containing “ultraconserved elements,” which are at least 200 base pairs in length and are 100 percent identical in the genomes of humans, mice and rats. The team used the gene-editing tool CRISPR-CAS9 to investigate enhancers near the Arx gene that, when defective, causes neurological and sexual-development disorders in mice and humans.
They aimed to determine whether there was some redundancy between the enhancer sequences and having two of them removed would compromise the viability or fertility of mice. Alternatively, perhaps the mice that seem normal when just one enhancer sequence was removed have a subtle difference. All mice were viable and fertile, and they teamed up with researchers at University of California, San Francisco (UCSF) to examine the morphological differences found in a part of the mouse forebrain that occurred when just one enhancer was deleted. 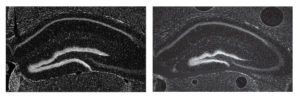 Pictured here in images captured by Athena Ypsilanti, a postdoctoral scholar at UCSF, is the mouse forebrain (left) compared with a mouse missing one ultraconserved enhancer (right).
Pictured here in images captured by Athena Ypsilanti, a postdoctoral scholar at UCSF, is the mouse forebrain (left) compared with a mouse missing one ultraconserved enhancer (right).
The abnormalities observed upon deletion of individual ultraconserved enhancers raise the question if deletion of less well-conserved enhancers generally causes similar problems. Are defects the exception or the rule? This question was investigated in a second study, led by postdoctoral researcher Marco Osterwalder, which focused on enhancers for limbs. The team, which also included Dickel, Visel, and Pennacchio, deleted ten limb enhancers near genes essential for limb development. They expected to see some anomalies but all ten mouse lines had perfectly normal-looking limbs. However, the team also observed some genes with two enhancers that appeared to be active at the same time during limb development.
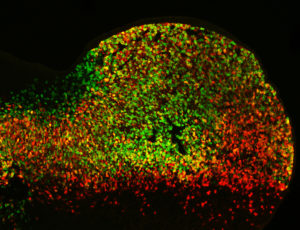
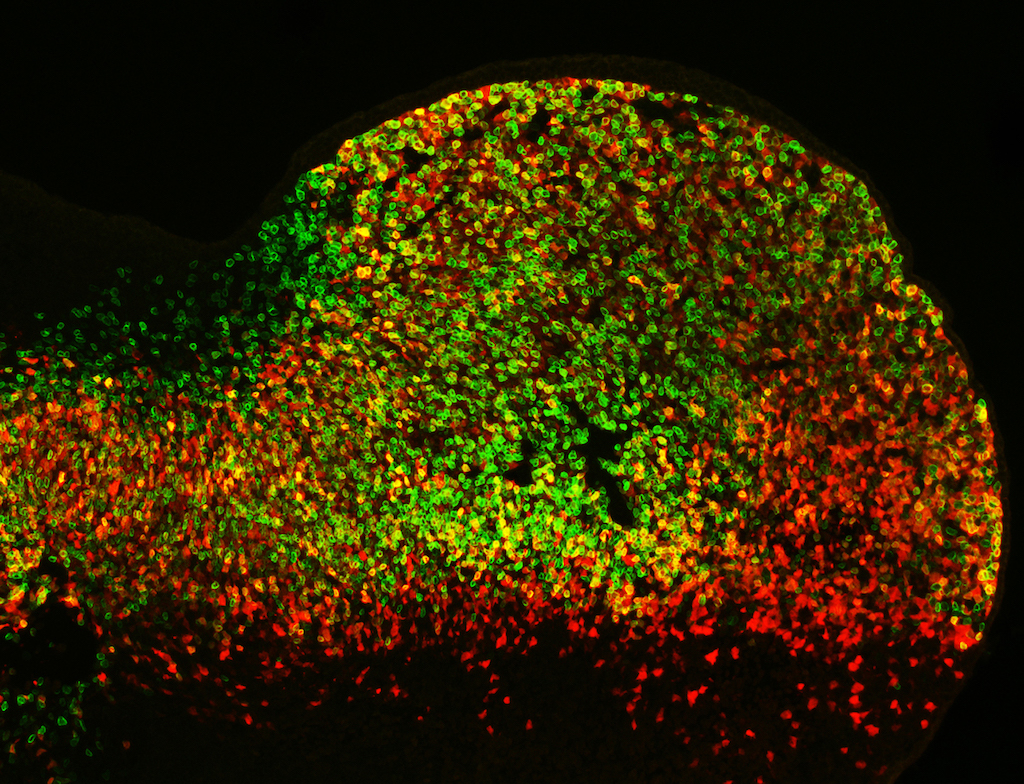
Cell activity patterns of two gene enhancers (red and green cells). Cells in which both enhancers are active appear yellow. (Credit: Marco Osterwalder).
When the team knocked out such pairs of limb enhancers with similar activity, they saw characteristics indicating that these enhancers functioned redundantly. To determine whether or not it’s common to have such a regulatory back-up system, the team computationally analyzed genomic datasets from many different tissues. They found that genes that control central processes in embryonic development are commonly equipped with sets of enhancers that are likely redundant. Read the Nature news piece that describes the Cell study. For a full description of the results, please read the Berkeley Lab News Center press release.