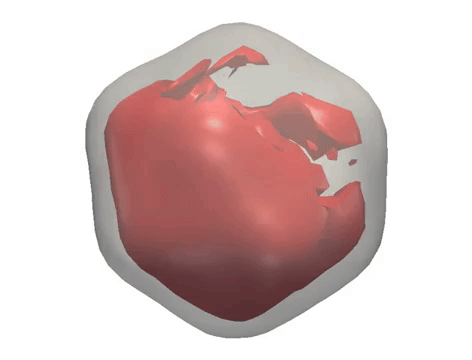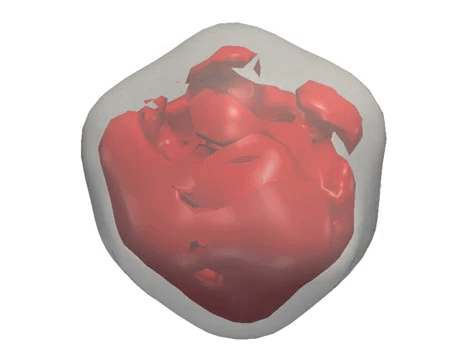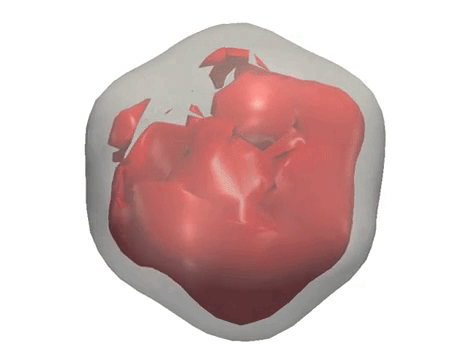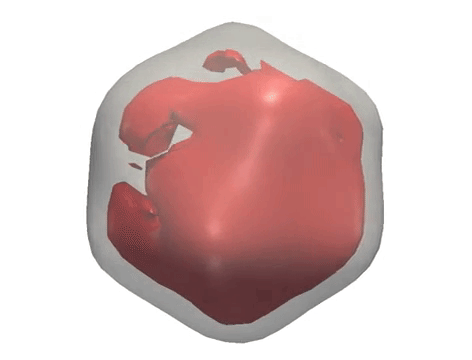
X-ray diffraction snapshots recorded in the XFEL experiment are reduced to a set of correlation data, which act as a ‘fingerprint’ of the 3D structure of a particle. Algorithms including M-TIP are then used to reconstruct the 3D structure of the particle. (Credit: European XFEL)
As part of an international team, researchers with Berkeley Lab’s Center for Advanced Mathematics for Energy Research Applications (CAMERA) employed their multi-tiered iterative phasing (M-TIP) algorithm to process X-ray free laser (XFEL) data taken from single virus particles and resolve their nanometer-scale structures in 3D. The new approach circumvents several challenges of imaging biomolecules that do not crystallize well, such as the random orientations of particles in solution and the asymmetrical structures of many viruses and proteins. Jeff Donatelli of the Computational Research Division’s Mathematics Group and Peter Zwart and Kanupriya Pande of the Biosciences Area’s Molecular Biophysics and Integrated Bioimaging (MBIB) division contributed to the work, the results of which were published in Physical Review Letters. Read more from the Berkeley Lab News Center.