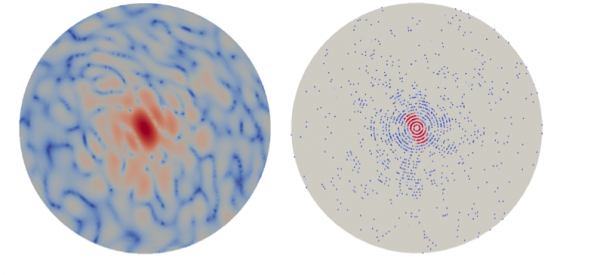
A cross-disciplinary effort by Berkeley Lab scientists has yielded a new algorithmic approach for determining 3D molecular structures from single-particle X-ray free-electron laser (XFEL) imaging. Peter Zwart of the Molecular Biophysics & Integrated Bioimaging Division (MBIB) worked with James Sethian and Jeffrey Donatelli of the Computational Research Division’s Mathematics Group to create the multi-tiered iterative phasing (M-TIP) framework, which uses advanced mathematical techniques to extract nano-scale biological structures from sparse and noisy diffraction data. A paper detailing the approach was published in the Proceedings of the National Academy of Sciences.
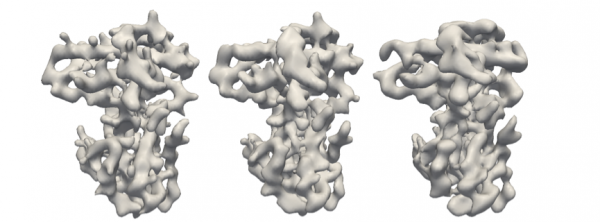
Original retinoblastoma protein (left) and reconstructions using the M-TIP algorithm with 24 clean images (middle) and 192 noisy images (right).
Zwart, Sethian, and Donatelli are all members of the the Center for Advanced Mathematics for Energy Research Applications (CAMERA). Jointly funded by the Advanced Scientific Computing Research and Basic Energy Sciences programs in Department of Energy’s Office of Science, CAMERA aims to develop the mathematics needed to process data from many of DOE’s advanced scientific facilities. The M-TIP project is part of a new collaborative initiative among CAMERA, SLAC National Accelerator Laboratory, the National Energy Research Scientific Computing Center (NERSC), and Los Alamos National Laboratory as part of DOE’s Exascale Computing Project (ECP). Read more from Berkeley Lab Computing Sciences.