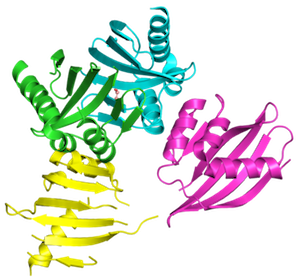
 Curved beta sheets are important for the architecture of protein cavities, such as enzyme active sites and ligand-binding pockets. Beginning by analyzing classic protein formations and running folding simulations, University of Washington (UW) researchers under the leadership of David Baker designed six protein folds inspired by naturally occurring protein superfamilies. A research report published in the January 13 issue of Science describes how scientists from multiple institutions compared the predicted models to physical structures of these designed proteins.
Curved beta sheets are important for the architecture of protein cavities, such as enzyme active sites and ligand-binding pockets. Beginning by analyzing classic protein formations and running folding simulations, University of Washington (UW) researchers under the leadership of David Baker designed six protein folds inspired by naturally occurring protein superfamilies. A research report published in the January 13 issue of Science describes how scientists from multiple institutions compared the predicted models to physical structures of these designed proteins.
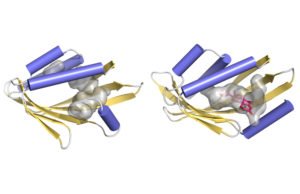
Illustration of how a curved protein surface (yellow-gold) forms a pocket (gray) to potentially fit molecules. At left is a schematic based on atomic-resolution data, at right is a hypothetical model. (Benjamin Basanta, Institute for Protein Design)
This interdisciplinary team included Jose Henrique Pereira, Banumathi Sankaran, and Peter Zwart of the Molecular Biophysics & Integrated Bioimaging Division, who collected X-ray data and refined the crystal structures. The collaboration is part of the Collaborative Crystallography program of the Berkeley Center for Structural Biology at the Advanced Light Source. All of the structures closely matched the predicted models, validating this method of designing the protein backbone to form cavities that could be customized to perform a reaction or facilitate a function of interest. Read more about the project on the UW Health Sciences NewsBeat and in this GeekWire article.