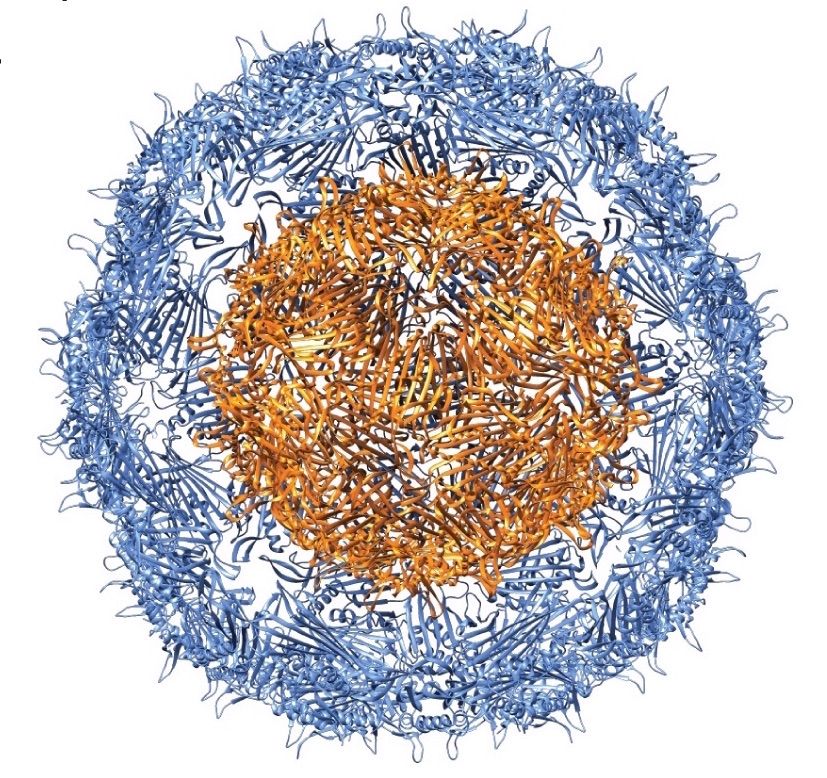
Viruses expertly package biological molecules and invade cells, two desirable features for introducing and delivering therapeutics, imaging agents, and other molecules to the body. Scientists have developed virus-like particles (VLPs) to encapsulate biologically active molecules in order to enhance their function. The drawback is in the limited ability to specifically produce appropriately sized containers for different applications.
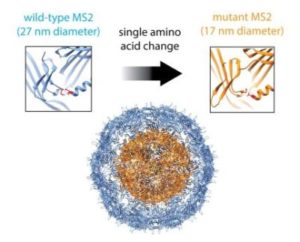 In their Nano Letters paper released August 23, 2016, a research team led by Danielle Tullman-Ercek, biologist faculty scientist in Molecular Biophysics & Integrated Bioimaging, described a new size selection method for virus-like particle assembly using chromatography. Working with Banumathi Sankaran and Peter Zwart in the Berkeley Center for Structural Biology at the Advanced Light Source, the scientists showed how a single amino acid mutation in the coat protein of bacteriophage MS2 causes a massive change in viral capsid shape (T3 to T1 icosahedral symmetry), which causes a difference in the size of the particle. The basic structural driver is a cis to trans peptide conformational change that results in the loss of a hydrogen bond. This slight modification of a binding interface ultimately results in a large change in the quaternary structure of the VLP assembly. This work has important implications for virus evolution theory, multi-protein assembly behavior, and protein-based nanomaterial development.
In their Nano Letters paper released August 23, 2016, a research team led by Danielle Tullman-Ercek, biologist faculty scientist in Molecular Biophysics & Integrated Bioimaging, described a new size selection method for virus-like particle assembly using chromatography. Working with Banumathi Sankaran and Peter Zwart in the Berkeley Center for Structural Biology at the Advanced Light Source, the scientists showed how a single amino acid mutation in the coat protein of bacteriophage MS2 causes a massive change in viral capsid shape (T3 to T1 icosahedral symmetry), which causes a difference in the size of the particle. The basic structural driver is a cis to trans peptide conformational change that results in the loss of a hydrogen bond. This slight modification of a binding interface ultimately results in a large change in the quaternary structure of the VLP assembly. This work has important implications for virus evolution theory, multi-protein assembly behavior, and protein-based nanomaterial development.