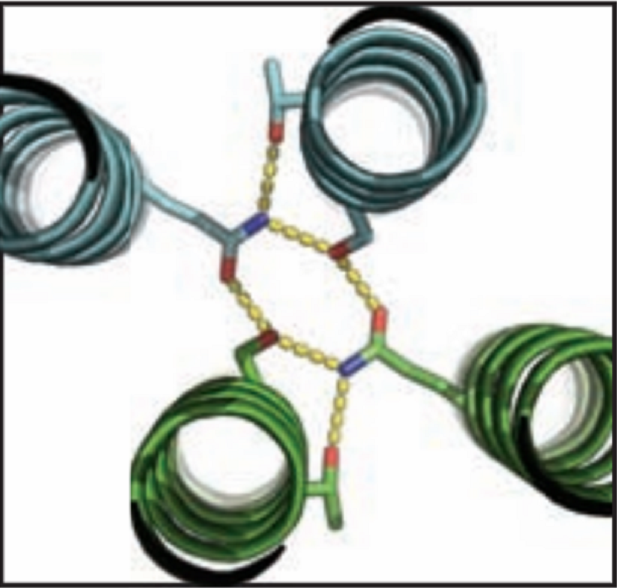
 Over the course of billions of years, nature has evolved particular molecular structures that form the basis of life, such as those found in nucleic acids and proteins. Using the natural form as a springboard, University of Washington researchers have designed protein homo-oligomers, or identical interacting subunits, which can contain interchangeable hydrogen bonding modules for building different structures or functions. The team of researchers, led by David Baker at the University of Washington, included Jose Henrique Pereira, Banumathi Sankaran, and Peter Zwart of the Molecular Biophysics & Integrated Bioimaging Division (MBIB).
Over the course of billions of years, nature has evolved particular molecular structures that form the basis of life, such as those found in nucleic acids and proteins. Using the natural form as a springboard, University of Washington researchers have designed protein homo-oligomers, or identical interacting subunits, which can contain interchangeable hydrogen bonding modules for building different structures or functions. The team of researchers, led by David Baker at the University of Washington, included Jose Henrique Pereira, Banumathi Sankaran, and Peter Zwart of the Molecular Biophysics & Integrated Bioimaging Division (MBIB).
Zwart runs two beamlines (8.2.1 and 8.2.2) that are part of the Berkeley Center Structural Biology in the Advanced Light Source and funded by the Howard Hughes Medical Institute for use by their investigators.  As a member of this elite group, Baker approached Zwart to collaborate. “David does great science, so I was all ears,” Zwart said. Baker told him that his group had about 100 molecules that had been expressed from synthetic genes in bacteria, all based on models predicted by their new computational algorithm, HBNet. Of these, 10 representative molecules were selected for X-ray crystallographic structure determination. Zwart has been working with Pereira and Sankaran on a novel crystallization and data collection service over the past couple of years and thought this would be a great opportunity to demonstrate its capabilities. “We have HHMI beamlines, a high-throughput method for crystallization, and three scientists who like to do science; they have a fantastic new program that demonstrates a leap forward in computer-aided protein design.” Zwart continued, “It was a match made in heaven.” Once the data were collected, the structures were determined using the Phenix software suite developed under the direction of Paul Adams, MBIB Division Director.
As a member of this elite group, Baker approached Zwart to collaborate. “David does great science, so I was all ears,” Zwart said. Baker told him that his group had about 100 molecules that had been expressed from synthetic genes in bacteria, all based on models predicted by their new computational algorithm, HBNet. Of these, 10 representative molecules were selected for X-ray crystallographic structure determination. Zwart has been working with Pereira and Sankaran on a novel crystallization and data collection service over the past couple of years and thought this would be a great opportunity to demonstrate its capabilities. “We have HHMI beamlines, a high-throughput method for crystallization, and three scientists who like to do science; they have a fantastic new program that demonstrates a leap forward in computer-aided protein design.” Zwart continued, “It was a match made in heaven.” Once the data were collected, the structures were determined using the Phenix software suite developed under the direction of Paul Adams, MBIB Division Director.
Those designs not chosen for X-ray crystallography were studied using small angle X-ray scattering (SAXS) on the SIBYLS beamline, where MBIB scientists Kathryn Burnett and Greg Hura collected the data. X-ray  crystallography showed that the structures and hydrogen-bond networks within the proteins were found to be in good agreement with the predicted model. SAXS provided further validation of 5 of these structures, as well as fundamental information about the degree of agreement between the proteins and models for the remaining predicted formations. “Most exciting,“ says Sankaran, “are the possibilities that arise from the ability to design synthetic molecules that combine the specificity of DNA–like binding with protein function.” Pereira agrees, adding, “This opens up huge opportunities for the fields of synthetic biology and materials science.”
crystallography showed that the structures and hydrogen-bond networks within the proteins were found to be in good agreement with the predicted model. SAXS provided further validation of 5 of these structures, as well as fundamental information about the degree of agreement between the proteins and models for the remaining predicted formations. “Most exciting,“ says Sankaran, “are the possibilities that arise from the ability to design synthetic molecules that combine the specificity of DNA–like binding with protein function.” Pereira agrees, adding, “This opens up huge opportunities for the fields of synthetic biology and materials science.”
A description of HBNet and subsequent validation of this algorithm were recently published in Science and the work was highlighted in a Science Insight. HBNet was also covered by Geekwire.