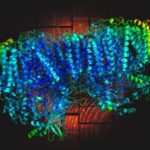
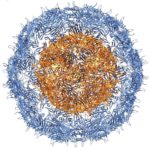
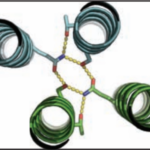
 University of Washington (UW) researchers have designed a novel protein with properties that could lead to the generation of new photoactive proteins. This three-fold symmetric, self-assembling protein homotrimer contains a highly stable noncanonical amino acid. Noncanonical amino acids are not found among the 20 encoded amino acids in the body and can contain modifications to allow for new functionality. In this case, this amino acid contains a bipyridine group that chelates metal, thereby introducing new photochemical properties into the protein interface, and nucleating the formation of the homotrimer.
University of Washington (UW) researchers have designed a novel protein with properties that could lead to the generation of new photoactive proteins. This three-fold symmetric, self-assembling protein homotrimer contains a highly stable noncanonical amino acid. Noncanonical amino acids are not found among the 20 encoded amino acids in the body and can contain modifications to allow for new functionality. In this case, this amino acid contains a bipyridine group that chelates metal, thereby introducing new photochemical properties into the protein interface, and nucleating the formation of the homotrimer.
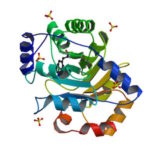
An article published last month in PNAS describes this work from a team of scientists led by David Baker at UW, which included Jose Henrique Pereira, Banumathi Sankaran, and Peter Zwart of the Molecular Biophysics & Integrated Bioimaging Division (MBIB). The MBIB scientists developed the crystal screen that was used to crystallize the novel protein and performed X-ray crystallography on Beamline 8.2.1 in the Berkeley Center for Structural Biology at the Advanced Light Source. Their X-ray crystallographic analysis of the homotrimer showed that the design process had near-atomic-level accuracy, demonstrating that computational protein design together with the utilization of noncanonical amino acids could be used to generate novel protein functions. These methods could be used to develop new therapeutics, biomaterials, and metalloproteins with useful optical or photochemical properties.